

| Basic Information | |
|---|---|
| Species | Panicum virgatum |
| Cazyme ID | Pavirv00049024m |
| Family | AA1 |
| Protein Properties | Length: 544 Molecular Weight: 59121.6 Isoelectric Point: 6.1091 |
| Chromosome | Chromosome/Scaffold: 072605 Start: 264 End: 2930 |
| Description | laccase 7 |
| View CDS | |
| External Links |
|---|
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| AA1 | 5 | 527 | 0 |
| SISQLCQPARIITAVNGQLPGPTIEAREGDTVVVHLVNQSPYNMTIHWHGVFQRGSPWEDGPSMVTQCPVQPGANYTYRFNVTDQEGTLWWHAHISFHRA TVYGALLIRPRGGAAAYPFSPKPHREETVILGEWWNASVYDLERMAFLTGNPAGPADAYTINGKPGDSDNCSTANQTYTFRVRRNETYLLRIINAALNTP MFLKVANHSFTVVAADAAYTTPYETDVVVVAPGQTVDALMAAGSAAGRYYMEASPYDSAVGATPPFNPTTATAVVEYAGAPAGGEPAKPPRPPSNDTATA FRFLSNLIALVLPGKPTVPLAVDTRMFVTVGLGVSDCQPDQLFCNRSRTVFSSSMNNASFVLPALGKPSMLEAHYRNESADVYTRDFPDRPPLAFDYTAD ASNTATLQYTTKSTKVRTLRYNETVEMVLQNTRLIAKESHPMHLHGFNFFVLAQGFGNYDEAAAAPGFNLVNPQERNTVAVPTGGWAVIRFVANNPGMWF MHCHFDAHLDLGLGMVFEVQDGP | |||
| Full Sequence |
|---|
| Protein Sequence Length: 544 Download |
| VGNLSISQLC QPARIITAVN GQLPGPTIEA REGDTVVVHL VNQSPYNMTI HWHGVFQRGS 60 PWEDGPSMVT QCPVQPGANY TYRFNVTDQE GTLWWHAHIS FHRATVYGAL LIRPRGGAAA 120 YPFSPKPHRE ETVILGEWWN ASVYDLERMA FLTGNPAGPA DAYTINGKPG DSDNCSTANQ 180 TYTFRVRRNE TYLLRIINAA LNTPMFLKVA NHSFTVVAAD AAYTTPYETD VVVVAPGQTV 240 DALMAAGSAA GRYYMEASPY DSAVGATPPF NPTTATAVVE YAGAPAGGEP AKPPRPPSND 300 TATAFRFLSN LIALVLPGKP TVPLAVDTRM FVTVGLGVSD CQPDQLFCNR SRTVFSSSMN 360 NASFVLPALG KPSMLEAHYR NESADVYTRD FPDRPPLAFD YTADASNTAT LQYTTKSTKV 420 RTLRYNETVE MVLQNTRLIA KESHPMHLHG FNFFVLAQGF GNYDEAAAAP GFNLVNPQER 480 NTVAVPTGGW AVIRFVANNP GMWFMHCHFD AHLDLGLGMV FEVQDGPTAE TSVPPPPLDL 540 PQC* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam07731 | Cu-oxidase_2 | 3.0e-42 | 390 | 527 | 141 | + Multicopper oxidase. This entry contains many divergent copper oxidase-like domains that are not recognised by the pfam00394 model. | ||
| pfam07732 | Cu-oxidase_3 | 5.0e-51 | 1 | 115 | 117 | + Multicopper oxidase. This entry contains many divergent copper oxidase-like domains that are not recognised by the pfam00394 model. | ||
| TIGR03388 | ascorbase | 5.0e-85 | 14 | 522 | 549 | + L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. | ||
| TIGR03389 | laccase | 0 | 5 | 543 | 540 | + laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005507 | copper ion binding |
| GO:0016491 | oxidoreductase activity |
| GO:0055114 | oxidation-reduction process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | ABA97328.1 | 0 | 1 | 543 | 112 | 657 | laccase family protein, putative [Oryza sativa (japonica cultivar-group)] |
| GenBank | EEC78752.1 | 0 | 1 | 543 | 31 | 576 | hypothetical protein OsI_18967 [Oryza sativa Indica Group] |
| GenBank | EEE62822.1 | 0 | 1 | 543 | 31 | 543 | hypothetical protein OsJ_17625 [Oryza sativa Japonica Group] |
| RefSeq | NP_001066518.1 | 0 | 1 | 543 | 31 | 576 | Os12g0259800 [Oryza sativa (japonica cultivar-group)] |
| RefSeq | XP_002442097.1 | 0 | 1 | 543 | 34 | 576 | hypothetical protein SORBIDRAFT_08g011530 [Sorghum bicolor] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1asq_B | 0 | 15 | 521 | 23 | 521 | A Chain A, The Three-Dimensional Structures Of Two Plant Beta-Glucan Endohydrolases With Distinct Substrate Specificities |
| PDB | 1asq_A | 0 | 15 | 521 | 23 | 521 | A Chain A, The Three-Dimensional Structures Of Two Plant Beta-Glucan Endohydrolases With Distinct Substrate Specificities |
| PDB | 1asp_B | 0 | 15 | 521 | 23 | 521 | A Chain A, The Three-Dimensional Structures Of Two Plant Beta-Glucan Endohydrolases With Distinct Substrate Specificities |
| PDB | 1asp_A | 0 | 15 | 521 | 23 | 521 | A Chain A, The Three-Dimensional Structures Of Two Plant Beta-Glucan Endohydrolases With Distinct Substrate Specificities |
| PDB | 1aso_B | 0 | 15 | 521 | 23 | 521 | A Chain A, The Three-Dimensional Structures Of Two Plant Beta-Glucan Endohydrolases With Distinct Substrate Specificities |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
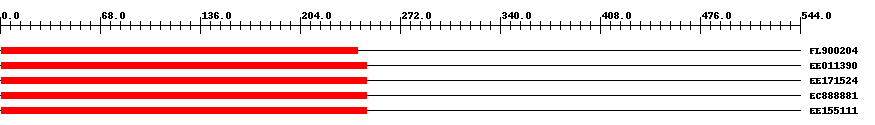 | ||||
| Hit | Length | Start | End | EValue |
| FL900204 | 244 | 1 | 243 | 0 |
| EE011390 | 249 | 1 | 249 | 0 |
| EE171524 | 249 | 1 | 249 | 0 |
| EC888881 | 249 | 1 | 249 | 0 |
| EE155111 | 249 | 1 | 249 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |