

| Basic Information | |
|---|---|
| Species | Arabidopsis thaliana |
| Cazyme ID | AT1G09010.1 |
| Family | GH2 |
| Protein Properties | Length: 945 Molecular Weight: 107661 Isoelectric Point: 6.4171 |
| Chromosome | Chromosome/Scaffold: 1 Start: 2895166 End: 2899384 |
| Description | mannosylglycoprotein endo-beta-mannosidase, putative, expressed |
| View CDS | |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH2 | 38 | 730 | 0 |
| SESRWMEAAVPGTVLGTLVKNKAIPDPFYGLENEAITDIADSGRDYYTFWFFTKFQCQRLLNQYVHLNFRAINYSAQVFVNGHKTELPKGMFRRHTLDVT DILHPESNLLALIVHPPDHPGTIPPEGGQGGDHEIGKDVAAQYVQGWDWICPIRDRNTGIWDEVSISVTGPVRIIDPHLVSTFFDDYKRAYLHVTAELEN KSTWNTECSVNIQITAELENGVCLVEHLQTENVLIPAQGRIQHTFKPLYFYKPELWWPNGMGKQNLYDILITVVVNEFGESDSWMQPFGFRKIESVIDSV TGGRLFKINGEPIFIRGGNWILSDGLLRLSKERYRTDIKFHADMNMNMIRCWGGGLAERPEFYHFCDIYGLLVWQEFWITGDVDGRGVPVSNPNGPLDHE LFLLCARDTVKLLRNHPSLALWVGGNEQVPPKDINEALKQDLRLHSYFETQLLSDKDSDPSVYLDGTRVYIQGSMWDGFADGKGNFTDGPYEIQYPEDFF KDTYYKYGFNPEVGSVGMPVAETIRATMPPEGWTIPLFKKGLDGFIKEVPNRMWDYHKYIPYSNPGKVHDQILMYGTPENLDDFCLKAQLVNYIQYRALF EGWSSQMWTKYTGVLIWKNQNPWTGLRGQFYDHLLDQTASFYGCRSAAEPVHVQLNLASYFVEVVNTTSKELSDVAIEASVWDLDGNCPYYKV | |||
| Full Sequence |
|---|
| Protein Sequence Length: 945 Download |
| MAEIGKTVLD FGWIAARSTE VDVNGVQLTT TNPPAISSES RWMEAAVPGT VLGTLVKNKA 60 IPDPFYGLEN EAITDIADSG RDYYTFWFFT KFQCQRLLNQ YVHLNFRAIN YSAQVFVNGH 120 KTELPKGMFR RHTLDVTDIL HPESNLLALI VHPPDHPGTI PPEGGQGGDH EIGKDVAAQY 180 VQGWDWICPI RDRNTGIWDE VSISVTGPVR IIDPHLVSTF FDDYKRAYLH VTAELENKST 240 WNTECSVNIQ ITAELENGVC LVEHLQTENV LIPAQGRIQH TFKPLYFYKP ELWWPNGMGK 300 QNLYDILITV VVNEFGESDS WMQPFGFRKI ESVIDSVTGG RLFKINGEPI FIRGGNWILS 360 DGLLRLSKER YRTDIKFHAD MNMNMIRCWG GGLAERPEFY HFCDIYGLLV WQEFWITGDV 420 DGRGVPVSNP NGPLDHELFL LCARDTVKLL RNHPSLALWV GGNEQVPPKD INEALKQDLR 480 LHSYFETQLL SDKDSDPSVY LDGTRVYIQG SMWDGFADGK GNFTDGPYEI QYPEDFFKDT 540 YYKYGFNPEV GSVGMPVAET IRATMPPEGW TIPLFKKGLD GFIKEVPNRM WDYHKYIPYS 600 NPGKVHDQIL MYGTPENLDD FCLKAQLVNY IQYRALFEGW SSQMWTKYTG VLIWKNQNPW 660 TGLRGQFYDH LLDQTASFYG CRSAAEPVHV QLNLASYFVE VVNTTSKELS DVAIEASVWD 720 LDGNCPYYKV FKIVSAPPKK VVKISEFKYP KAANPKHVYF LLLKLYTVSD KAVISRNFYW 780 LHLPGKNYTL LEPYRKKQIP LKITCNAVMV GSRYELEVNV HNTSRANLAK NVVQEDEKRD 840 LGLLQKLFSR CVVSADSNRG LKVVEMKGSD SGVAFFLRFS VHNAETEKQD TRILPVHYSD 900 NYFSLVPGES MSFKISFAAP TGMKKSPRVM LQGWNYPDRF SVFG* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
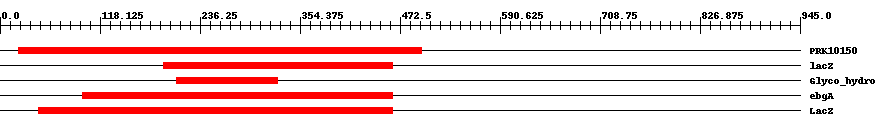 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PRK10150 | PRK10150 | 3.0e-7 | 22 | 498 | 510 | + beta-D-glucuronidase; Provisional | ||
| PRK09525 | lacZ | 5.0e-8 | 193 | 464 | 295 | + beta-D-galactosidase; Reviewed | ||
| pfam00703 | Glyco_hydro_2 | 4.0e-11 | 209 | 328 | 120 | + Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. | ||
| PRK10340 | ebgA | 6.0e-16 | 98 | 464 | 378 | + cryptic beta-D-galactosidase subunit alpha; Reviewed | ||
| COG3250 | LacZ | 1.0e-40 | 46 | 464 | 427 | + Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism] | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0003824 | catalytic activity |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0005773 | vacuole |
| GO:0005975 | carbohydrate metabolic process |
| GO:0043169 | cation binding |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAB70407.1 | 0.00007 | 1 | 26 | 1 | 26 | Contains similarity to Bos beta-mannosidase (gb |
| GenBank | AAB70407.1 | 0 | 90 | 944 | 19 | 810 | Contains similarity to Bos beta-mannosidase (gb |
| DDBJ | BAE97370.1 | 0 | 1 | 944 | 1 | 946 | endo-beta-mannosidase [Brassica oleracea] |
| RefSeq | NP_563833.2 | 0 | 1 | 944 | 1 | 944 | glycoside hydrolase family 2 protein [Arabidopsis thaliana] |
| RefSeq | XP_002319539.1 | 0 | 1 | 943 | 1 | 973 | predicted protein [Populus trichocarpa] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2vzo_B | 0 | 40 | 781 | 79 | 767 | A Chain A, Crystal Structure Of Amycolatopsis Orientalis Exo- Chitosanase Csxa |
| PDB | 2vzo_A | 0 | 40 | 781 | 79 | 767 | A Chain A, Crystal Structure Of Amycolatopsis Orientalis Exo- Chitosanase Csxa |
| PDB | 2x09_B | 0 | 40 | 781 | 79 | 767 | A Chain A, Crystal Structure Of Amycolatopsis Orientalis Exo- Chitosanase Csxa |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
 | ||||
| Hit | Length | Start | End | EValue |
| ES793920 | 376 | 372 | 740 | 0 |
| GO871600 | 333 | 300 | 624 | 0 |
| EX109426 | 264 | 104 | 366 | 0 |
| GO847639 | 325 | 509 | 833 | 0 |
| ES832790 | 305 | 490 | 794 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |