

| Basic Information | |
|---|---|
| Species | Glycine max |
| Cazyme ID | Glyma10g29550.1 |
| Family | GH2 |
| Protein Properties | Length: 976 Molecular Weight: 110474 Isoelectric Point: 6.5079 |
| Chromosome | Chromosome/Scaffold: 10 Start: 38393119 End: 38399083 |
| Description | glycoside hydrolase family 2 protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH2 | 32 | 756 | 0 |
| PSASTQPWMEAIVPGTVLATLVKNKAVPDPFYGLGNEAILDIADSGREYYTFWFFTTFQCKLSGNQHCDLNFRGINYSADVYLNGHEMVLPKGMFRRHSL DVTNIIHSDSSNLLAVLVHPPDHPGSIPPQGGQGGDHEIGKDVATQYVQGWDWMAPIRDRNTGIWDEVSISITGPVKIIDPHLVSSFFDNYKRVYLHATT ELENRSSLIAECSLSIHVTTELEGNIHLVEQLQTQNLSIPARSRVQYTFPELFFYKPNLWWPNGMGKQSLYNVIITIDVKGHGESDSWNHHFGFRKIESH IDVATGGRLFKVNGEPIFIRGGNWILSDGLLRLSKKRYETDIKFHADMNFNMIRCWGGGLAERPEFYRCCDYYGLLVWQEFWITGDVDGRGIPVSNPNGP LDHDLFLFCARDTVKLLRNHPSLALWVGGNEQTPPDDLNVALKNDLKLHPYFGHAEEKEKPVGDLSPRLGDPSQYLDGTRIYIQGSLWDGFADGEGDFSD GPYEIQNPEDFFTDSFYNHGFNPEVGSVGMPVASTIRAIMPSEGWQIPLFNKLPNGYVEEVPNPIWKYHKYIPYSNPTKKVHDQIQLYGDVKDLDDFCLK AQLVNYIQYRALLEGWTSRMWKKYTGVLIWKTQNPWTGLRGQFYDHLLDQTAGFYGCRCAAEPIHVQLNLATYFIEVVNTTSEELSNLAMEVSVWDLEGT CPHYKVHENLTALPKKVTPIVDMKY | |||
| Full Sequence |
|---|
| Protein Sequence Length: 976 Download |
| MMAKTTLDSG WLAARSTEVH FTGTQLTTTH PPSASTQPWM EAIVPGTVLA TLVKNKAVPD 60 PFYGLGNEAI LDIADSGREY YTFWFFTTFQ CKLSGNQHCD LNFRGINYSA DVYLNGHEMV 120 LPKGMFRRHS LDVTNIIHSD SSNLLAVLVH PPDHPGSIPP QGGQGGDHEI GKDVATQYVQ 180 GWDWMAPIRD RNTGIWDEVS ISITGPVKII DPHLVSSFFD NYKRVYLHAT TELENRSSLI 240 AECSLSIHVT TELEGNIHLV EQLQTQNLSI PARSRVQYTF PELFFYKPNL WWPNGMGKQS 300 LYNVIITIDV KGHGESDSWN HHFGFRKIES HIDVATGGRL FKVNGEPIFI RGGNWILSDG 360 LLRLSKKRYE TDIKFHADMN FNMIRCWGGG LAERPEFYRC CDYYGLLVWQ EFWITGDVDG 420 RGIPVSNPNG PLDHDLFLFC ARDTVKLLRN HPSLALWVGG NEQTPPDDLN VALKNDLKLH 480 PYFGHAEEKE KPVGDLSPRL GDPSQYLDGT RIYIQGSLWD GFADGEGDFS DGPYEIQNPE 540 DFFTDSFYNH GFNPEVGSVG MPVASTIRAI MPSEGWQIPL FNKLPNGYVE EVPNPIWKYH 600 KYIPYSNPTK KVHDQIQLYG DVKDLDDFCL KAQLVNYIQY RALLEGWTSR MWKKYTGVLI 660 WKTQNPWTGL RGQFYDHLLD QTAGFYGCRC AAEPIHVQLN LATYFIEVVN TTSEELSNLA 720 MEVSVWDLEG TCPHYKVHEN LTALPKKVTP IVDMKYLKSK NPKPVYFLLL KLYKMSDNTI 780 LSRNFYWLHL PGGDYKLLEP YRKKKIPLKI TSEVFIEGST YILQMHVQNT SKKPDSKSLT 840 MVHSSTARQS SGCFVTDSLE TVDSGTGKEH EVGWFKGIHK CFAGKSHGLK VSEINGQDTG 900 VAFFLHFSVH ASNKDHKEGE DTRILPIHYS DNYFSLVPGE TMTIKISFEV PSGVSPCVTL 960 RGWNYQGQTI HEALY* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam02836 | Glyco_hydro_2_C | 5.0e-5 | 334 | 462 | 139 | + Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. | ||
| PRK09525 | lacZ | 8.0e-7 | 288 | 462 | 202 | + beta-D-galactosidase; Reviewed | ||
| pfam00703 | Glyco_hydro_2 | 9.0e-11 | 207 | 326 | 120 | + Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. | ||
| PRK10340 | ebgA | 1.0e-11 | 96 | 462 | 382 | + cryptic beta-D-galactosidase subunit alpha; Reviewed | ||
| COG3250 | LacZ | 9.0e-39 | 44 | 462 | 443 | + Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism] | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0005975 | carbohydrate metabolic process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| EMBL | CAN82620.1 | 0 | 2 | 964 | 4 | 965 | hypothetical protein [Vitis vinifera] |
| EMBL | CBI36793.1 | 0 | 2 | 964 | 4 | 943 | unnamed protein product [Vitis vinifera] |
| RefSeq | XP_002284576.1 | 0 | 2 | 964 | 4 | 965 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002319539.1 | 0 | 2 | 966 | 4 | 967 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002512381.1 | 0 | 2 | 967 | 4 | 968 | beta-mannosidase, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2vzo_B | 0 | 32 | 788 | 74 | 767 | A Chain A, Crystal Structure Of Amycolatopsis Orientalis Exo- Chitosanase Csxa |
| PDB | 2vzo_A | 0 | 32 | 788 | 74 | 767 | A Chain A, Crystal Structure Of Amycolatopsis Orientalis Exo- Chitosanase Csxa |
| PDB | 2x09_B | 0 | 32 | 788 | 74 | 767 | A Chain A, Crystal Structure Of Amycolatopsis Orientalis Exo- Chitosanase Csxa |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
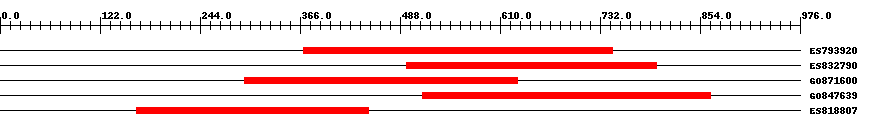 | ||||
| Hit | Length | Start | End | EValue |
| ES793920 | 383 | 370 | 747 | 0 |
| ES832790 | 306 | 496 | 801 | 0 |
| GO871600 | 334 | 298 | 631 | 0 |
| GO847639 | 353 | 515 | 867 | 0 |
| ES818807 | 283 | 167 | 449 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |