

| Basic Information | |
|---|---|
| Species | Panicum virgatum |
| Cazyme ID | Pavirv00063462m |
| Family | GT20 |
| Protein Properties | Length: 906 Molecular Weight: 101700 Isoelectric Point: 5.2935 |
| Chromosome | Chromosome/Scaffold: 022089 Start: 4410 End: 8530 |
| Description | trehalose-phosphatase/synthase 7 |
| View CDS | |
| External Links |
|---|
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GT20 | 73 | 572 | 0 |
| AVGERIIVVANQLPVVARRRPDGRGWVFSWDEDSLLLRLRDGVPDEMEVFFVGSLRADVPPAEQDEVSQTLIDGFRCAPVFLSPELNERFYHHFCKGYLW PLFHYMLPFAAQLSPTTEAAPSSDGGRFDRSAWEAYVLANKHFYEKVVEVINPEDDYIWVHDYHLMALPTFLRRRFNRLRIGFFLHSPFPSSEIYRTLPV REEVLRTLLNCDLIGFHTFDYARHFLSCCSRMLGLEYQSKRGYIGLEYFGRTVGIKIMPMGIHMGQLQSVLRLPEQEQKVAELRQRFEGKTVFLGVDDTD IFKGINLKLLAFETMLRMHPKWQGRAVLVQIANPPRGKGKELETIQAEIRVTCERINRDFGRTGYSPVVFIDRNVPSAERLAYYTIAECVVVTAVRDGMN LTPYEYIVCRQGIPGSESAPEVSGPKKSMLVVSEFIGCSPSLSGAIRVNPWNIETTAEALNEAISVSEQEKQLRHGKHYRYVSTHDVAYWSRSFVQDLER | |||
| Full Sequence |
|---|
| Protein Sequence Length: 906 Download |
| MMSRSYTNLL DLAAGNFAAL GPSGGGRRRS GSFGMKRMPR VMTVPGPLSD LDDDDDDEQA 60 ATSSVASDVP SSAVGERIIV VANQLPVVAR RRPDGRGWVF SWDEDSLLLR LRDGVPDEME 120 VFFVGSLRAD VPPAEQDEVS QTLIDGFRCA PVFLSPELNE RFYHHFCKGY LWPLFHYMLP 180 FAAQLSPTTE AAPSSDGGRF DRSAWEAYVL ANKHFYEKVV EVINPEDDYI WVHDYHLMAL 240 PTFLRRRFNR LRIGFFLHSP FPSSEIYRTL PVREEVLRTL LNCDLIGFHT FDYARHFLSC 300 CSRMLGLEYQ SKRGYIGLEY FGRTVGIKIM PMGIHMGQLQ SVLRLPEQEQ KVAELRQRFE 360 GKTVFLGVDD TDIFKGINLK LLAFETMLRM HPKWQGRAVL VQIANPPRGK GKELETIQAE 420 IRVTCERINR DFGRTGYSPV VFIDRNVPSA ERLAYYTIAE CVVVTAVRDG MNLTPYEYIV 480 CRQGIPGSES APEVSGPKKS MLVVSEFIGC SPSLSGAIRV NPWNIETTAE ALNEAISVSE 540 QEKQLRHGKH YRYVSTHDVA YWSRSFVQDL ERACKDHFRK PCWGIGLGFG FRVVALDPHF 600 SKLNLDTIVM SYERAKSRAI LLDYDGTLVP QASINKEPSA EIVSIINTLC SDSNNIVFIV 660 SGRSRDSLGS MFASCPKLGL AAEHGYFLRW TRDEEWQTIT RTPDFGWMQM AEPVMNLYTE 720 ATDGSYIETK ETALVWHHQD ADPGFGSSQA KEMLDHLESV LANEPVAVKS GQFIVEVKPQ 780 GVSKGLVAEK ILTLMKEKGQ QADFVLCIGD DRSDEDMFEN IADVMKRNIV APKTPLFACT 840 VGQKPSKARF YLDDTFEVVN MLSSLADASD PDPIIELEDD LATSVSSIDI SDEPPKFGNR 900 RDEGS* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN03063 | PLN03063 | 4.0e-172 | 77 | 870 | 836 | + alpha,alpha-trehalose-phosphate synthase (UDP-forming); Provisional | ||
| PLN02205 | PLN02205 | 0 | 1 | 870 | 880 | + alpha,alpha-trehalose-phosphate synthase [UDP-forming] | ||
| pfam00982 | Glyco_transf_20 | 0 | 76 | 572 | 501 | + Glycosyltransferase family 20. Members of this family belong to glycosyl transferase family 20. OtsA (Trehalose-6-phosphate synthase) is homologous to regions in the subunits of yeast trehalose-6-phosphate synthase/phosphate complex. | ||
| PRK14501 | PRK14501 | 0 | 77 | 868 | 803 | + putative bifunctional trehalose-6-phosphate synthase/HAD hydrolase subfamily IIB; Provisional | ||
| cd03788 | GT1_TPS | 0 | 77 | 571 | 495 | + Trehalose-6-Phosphate Synthase (TPS) is a glycosyltransferase that catalyses the synthesis of alpha,alpha-1,1-trehalose-6-phosphate from glucose-6-phosphate using a UDP-glucose donor. It is a key enzyme in the trehalose synthesis pathway. Trehalose is a nonreducing disaccharide present in a wide variety of organisms and may serve as a source of energy and carbon. It is characterized most notably in insect, plant, and microbial cells. Its production is often associated with a variety of stress conditions, including desiccation, dehydration, heat, cold, and oxidation. This family represents the catalytic domain of the TPS. Some members of this domain family coexist with a C-terminal trehalose phosphatase domain. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0003824 | catalytic activity |
| GO:0005992 | trehalose biosynthetic process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | EEC79533.1 | 0 | 1 | 905 | 1 | 899 | hypothetical protein OsI_20637 [Oryza sativa Indica Group] |
| GenBank | EEE64369.1 | 0 | 1 | 905 | 1 | 899 | hypothetical protein OsJ_19211 [Oryza sativa Japonica Group] |
| RefSeq | NP_001056050.1 | 0 | 1 | 905 | 1 | 899 | Os05g0517200 [Oryza sativa (japonica cultivar-group)] |
| RefSeq | XP_002440081.1 | 0 | 1 | 905 | 1 | 906 | hypothetical protein SORBIDRAFT_09g025660 [Sorghum bicolor] |
| RefSeq | XP_002458490.1 | 0 | 1 | 905 | 1 | 910 | hypothetical protein SORBIDRAFT_03g034640 [Sorghum bicolor] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2wtx_D | 0 | 154 | 572 | 68 | 453 | A Chain A, Insight Into The Mechanism Of Enzymatic Glycosyltransfer With Retention Through The Synthesis And Analysis Of Bisubstrate Glycomimetics Of Trehalose-6-Phosphate Synthase |
| PDB | 2wtx_C | 0 | 154 | 572 | 68 | 453 | A Chain A, Insight Into The Mechanism Of Enzymatic Glycosyltransfer With Retention Through The Synthesis And Analysis Of Bisubstrate Glycomimetics Of Trehalose-6-Phosphate Synthase |
| PDB | 2wtx_B | 0 | 154 | 572 | 68 | 453 | A Chain A, Insight Into The Mechanism Of Enzymatic Glycosyltransfer With Retention Through The Synthesis And Analysis Of Bisubstrate Glycomimetics Of Trehalose-6-Phosphate Synthase |
| PDB | 2wtx_A | 0 | 154 | 572 | 68 | 453 | A Chain A, Insight Into The Mechanism Of Enzymatic Glycosyltransfer With Retention Through The Synthesis And Analysis Of Bisubstrate Glycomimetics Of Trehalose-6-Phosphate Synthase |
| PDB | 1uqu_B | 0 | 154 | 572 | 68 | 453 | A Chain A, Trehalose-6-Phosphate From E. Coli Bound With Udp-Glucose. |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| trehalose biosynthesis I | TREHALOSE6PSYN-RXN | EC-2.4.1.15 | α,α-trehalose-phosphate synthase (UDP-forming) |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
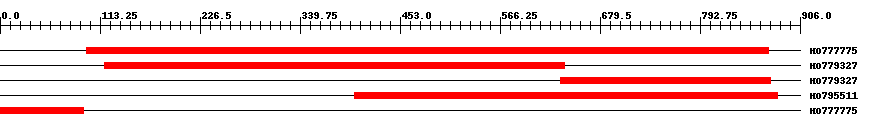 | ||||
| Hit | Length | Start | End | EValue |
| HO777775 | 783 | 98 | 870 | 0 |
| HO779327 | 524 | 118 | 639 | 0 |
| HO779327 | 243 | 635 | 873 | 0 |
| HO795511 | 479 | 402 | 880 | 0 |
| HO777775 | 94 | 1 | 94 | 0.29 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |