
| APIS family ID | APIS195 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inhibited defense system | toxin-antitoxin (TA) | |||||||||||||
| CLAN ID | CLAN065 | |||||||||||||
| Seed protein property |
|
|||||||||||||
| Phage property |
|
|||||||||||||
| PMID/References | PMID:39026772 | |||||||||||||
| PDB structures | ; | |||||||||||||
| Pfam domains | PF08010.16 | |||||||||||||
| Phrog | ||||||||||||||
| Host taxa | d__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacterales;f__Enterobacteriaceae;g__Escherichia; | |||||||||||||
| Gene Location | Start: ; End: ; Strand: | |||||||||||||
| Description | Anti-DarT factor NADAR (AdfN), removes ADP-ribose modifications from phage DNA during infection thereby enabling replication in DarTG1-containing bacteria. AdfN, like DarG1, is in the NADAR superfamily of ADP-ribosylglycohydrolases. | |||||||||||||
pTM is for the estimate of the TM-score, which is obtained from a pairwise error prediction. The higher pTM score indicates better model quality.
pLDDT is for per-residue accuracy of the structure, which representes the quality of the residue. A higher value indicates better prediction accuracy. More detail please see AlphaFold .
| Hit | Query start | Query end | Hit start | Hit end | TMscore(ali) | TMscore(avg) |
|---|---|---|---|---|---|---|
| MGYP000187068478.pdb | 1 | 152 | 1 | 152 | 0.9745 | 0.9674 |
| MGYP001571694117.pdb | 1 | 151 | 2 | 152 | 0.9433 | 0.9313 |
| MGYP000012094249.pdb | 4 | 152 | 1 | 146 | 0.9514 | 0.9252 |
| MGYP003621366742.pdb | 4 | 152 | 1 | 146 | 0.952 | 0.925 |
| MGYP002508153472.pdb | 4 | 149 | 1 | 144 | 0.9575 | 0.9218 |
| MGYP000669273966.pdb | 3 | 152 | 1 | 147 | 0.9483 | 0.9195 |
| MGYP003487091401.pdb | 4 | 152 | 1 | 149 | 0.9406 | 0.9192 |
| MGYP002558511419.pdb | 1 | 152 | 2 | 151 | 0.9333 | 0.916 |
| MGYP003308194965.pdb | 2 | 152 | 1 | 148 | 0.9433 | 0.9156 |
| MGYP000480180796.pdb | 3 | 152 | 1 | 150 | 0.9369 | 0.9154 |
| Fam ID | Seed protein | Member count | Model | Alignment | APIS195 | AdfN | 192 | HMM model | Member alignment |
|---|
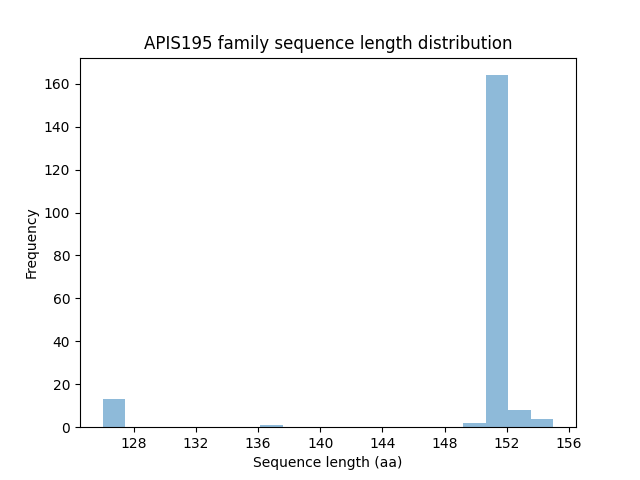
| Member | pident | evalue | bitscore | qcovs | phage | host | SourceDB | Pfam | Phrog |
|---|---|---|---|---|---|---|---|---|---|
| ON528737_00223 | 94.500 | 3.767E-96 | 304 | 1.000% | Shigella phage ESh27 | Shigella | INPHARED | PF08010 | phrog_ |
| MT682709_00071 | 97.700 | 3.984E-100 | 315 | 1.000% | Escherichia phage vB_EcoM_SP1 | Escherichia | INPHARED | PF08010 | phrog_ |
| OL741432_00206 | 97.500 | 7.490E-100 | 314 | 1.000% | Proteus phage Isf-Pm2 | Proteus | INPHARED | PF08010 | phrog_ |
| MGV-GENOME-0377573_182 | 95.700 | 1.169E-97 | 308 | 1.000% | MGV | PF08010 | phrog_652 | ||
| MGV-GENOME-0377451_188 | 96.600 | 9.358E-99 | 311 | 1.000% | MGV | PF08010 | phrog_ | ||
| OP557970_00115 | 97.300 | 1.027E-99 | 314 | 1.000% | Escherichia phage ph0021 | Escherichia | INPHARED | PF08010 | phrog_ |
| MZ311865_00117 | 95.700 | 1.169E-97 | 308 | 1.000% | Escherichia phage vB_EcoM_LNA2 | Escherichia | INPHARED | PF08010 | phrog_ |
| IMGVR_UViG_2974743505_000001|2974743505|2974743546 | 98.500 | 3.188E-101 | 318 | 1.000% | IMGVR | PF08010 | phrog_ | ||
| MW286157_00132 | 95.800 | 8.528E-98 | 308 | 1.000% | Escherichia phage vB_EcoM-BECP11 | Escherichia | INPHARED | PF08010 | phrog_ |
| AY303349_00211 | 96.200 | 8.682E-88 | 280 | 0.901% | Escherichia phage RB69 | Escherichia | INPHARED | PF08010 | phrog_652 |
| IMGVR_UViG_2706795326_000001|2706795326|2708651494 | 96.000 | 7.764E-80 | 257 | 0.829% | IMGVR | PF08010 | phrog_ | ||
| MW250787_00085 | 87.000 | 7.912E-87 | 277 | 1.000% | Escherichia phage UPEC07 | Escherichia | INPHARED | PF08010 | phrog_ |
| MZ234052_00167 | 97.800 | 2.905E-100 | 316 | 1.000% | Escherichia phage vB_EcoM-PM133 | Escherichia | INPHARED | PF08010 | phrog_652 |
| IMGVR_UViG_2706795356_000001|2706795356|2708653673 | 87.600 | 1.632E-87 | 279 | 1.000% | IMGVR | PF08010 | phrog_ | ||
| LR597660_00203 | 97.500 | 7.490E-100 | 314 | 1.000% | Escherichia phage T4_ev151 | Escherichia | INPHARED | PF08010 | phrog_ |
| IMGVR_UViG_3300010279_000005|3300010279|Ga0129313_1000058187 | 96.500 | 2.196E-80 | 258 | 0.829% | IMGVR | PF08010 | phrog_ | ||
| IMGVR_UViG_2706795401_000001|2706795401|2708657425 | 87.700 | 1.191E-87 | 279 | 1.000% | IMGVR | PF08010 | phrog_ | ||
| OQ349392_00068 | 96.600 | 9.358E-99 | 311 | 1.000% | Escherichia phage KIT06 | Escherichia | INPHARED | PF08010 | phrog_ |
| OK018184_00214 | 98.000 | 1.545E-100 | 316 | 1.000% | Shigella phage CT01 | Shigella | INPHARED | PF08010 | phrog_ |
| LR597657_00199 | 97.700 | 3.984E-100 | 315 | 1.000% | Escherichia phage T4_ev240 | Escherichia | INPHARED | PF08010 | phrog_ |
| KT184308_00112 | 97.100 | 1.931E-99 | 313 | 1.000% | Enterobacteria phage Aplg8 | Enterobacteria | INPHARED | PF08010 | phrog_ |
| KU867876_00209 | 96.600 | 9.358E-99 | 311 | 1.000% | Escherichia phage vB_EcoM-UFV13 | Escherichia | INPHARED | PF08010 | phrog_ |
| IMGVR_UViG_2706795408_000001|2706795408|2708658342 | 88.200 | 2.456E-88 | 281 | 1.000% | IMGVR | PF08010 | phrog_ | ||
| OP137232_00167 | 96.800 | 4.977E-99 | 312 | 1.000% | Shigella phage S2_01 | Shigella | INPHARED | PF08010 | phrog_652 |
| OP197928_00208 | 96.500 | 1.283E-98 | 311 | 1.000% | Shigella phage A2 | Shigella | INPHARED | PF08010 | phrog_ |
| OK272484_00207 | 97.600 | 5.462E-100 | 315 | 1.000% | Escherichia phage JLBYU22 | Escherichia | INPHARED | PF08010 | phrog_ |
| LR215724_00209 | 96.700 | 6.825E-99 | 312 | 1.000% | Yersinia phage fPS-65 | Yersinia | INPHARED | PF08010 | phrog_ |
| KP869105_00236 | 97.200 | 1.408E-99 | 314 | 1.000% | Escherichia coli O157 typing phage 7 | Escherichia | INPHARED | PF08010 | phrog_ |
| OM386666_00236 | 97.200 | 1.408E-99 | 314 | 1.000% | Escherichia phage vB_EcoM_ESCO58 | Escherichia | INPHARED | PF08010 | phrog_ |
| OM135583_00200 | 96.600 | 9.358E-99 | 311 | 1.000% | Escherichia phage EP01 | Escherichia | INPHARED | PF08010 | phrog_ |
| MZ753803_00205 | 97.900 | 2.119E-100 | 316 | 1.000% | Escherichia phage U115 | Escherichia | INPHARED | PF08010 | phrog_ |
| MF327009_00055 | 97.500 | 7.490E-100 | 314 | 1.000% | Shigella phage Sf25 | Shigella | INPHARED | PF08010 | phrog_ |
| MF158046_00049 | 96.500 | 1.283E-98 | 311 | 1.000% | Shigella phage Sf23 | Shigella | INPHARED | PF08010 | phrog_ |
| MGV-GENOME-0377569_48 | 97.800 | 2.905E-100 | 316 | 1.000% | MGV | PF08010 | phrog_ | ||
| OL870317_00210 | 97.700 | 3.984E-100 | 315 | 1.000% | Escherichia phage UTI-E4 | Escherichia | INPHARED | PF08010 | phrog_ |
| OL960581_00200 | 96.200 | 2.412E-98 | 310 | 1.000% | Escherichia phage vB_EcoM_SA20RB | Escherichia | INPHARED | PF08010 | phrog_ |
| OK272476_00208 | 97.700 | 3.984E-100 | 315 | 1.000% | Escherichia phage JLBYU24 | Escherichia | INPHARED | PF08010 | phrog_ |
| MZ502380_00204 | 96.600 | 9.358E-99 | 311 | 1.000% | Escherichia phage vB_EcoM_Nami | Escherichia | INPHARED | PF08010 | phrog_ |
| MW341595_00259 | 97.500 | 7.490E-100 | 314 | 1.000% | Shigella phage Sfk20 | Shigella | INPHARED | PF08010 | phrog_ |
| MT446420_00020 | 88.800 | 5.067E-89 | 283 | 1.000% | Escherichia virus TH54 | Escherichia | INPHARED | PF08010 | phrog_ |
| MK373787_00205 | 96.700 | 6.825E-99 | 312 | 1.000% | Escherichia phage vB_EcoM_G8 | Escherichia | INPHARED | PF08010 | phrog_ |
| LN881737_00231 | 97.000 | 2.647E-99 | 313 | 1.000% | Escherichia phage slur13 | Escherichia | INPHARED | PF08010 | phrog_ |
| IMGVR_UViG_3300029310_000117|3300029310|Ga0134545_100420315 | 95.600 | 1.603E-97 | 308 | 1.000% | IMGVR | PF08010 | phrog_ | ||
| KY703222_00054 | 95.900 | 6.219E-98 | 309 | 1.000% | Escherichia phage phiC120 | Escherichia | INPHARED | PF08010 | phrog_ |
| ON506924_00208 | 97.300 | 1.027E-99 | 314 | 1.000% | Escherichia phage vB_EcoM_SCS4 | Escherichia | INPHARED | PF08010 | phrog_ |
| OK040907_00202 | 97.800 | 2.905E-100 | 316 | 1.000% | Enterobacteria phage Ac3 | Enterobacteria | INPHARED | PF08010 | phrog_ |
| OL960577_00212 | 95.800 | 8.528E-98 | 308 | 1.000% | Escherichia phage vB_EcoM_SA35RD | Escherichia | INPHARED | PF08010 | phrog_ |
| MT446415_00278 | 88.500 | 1.306E-88 | 282 | 1.000% | Escherichia virus TH44 | Escherichia | INPHARED | PF08010 | phrog_ |
| LN881732_00058 | 98.000 | 1.545E-100 | 316 | 1.000% | Escherichia phage slur07 | Escherichia | INPHARED | PF08010 | phrog_ |
| JN986846_00209 | 98.200 | 8.219E-101 | 317 | 1.000% | Escherichia phage vB_EcoM_ACG-C40 | Escherichia | INPHARED | PF08010 | phrog_ |
| MGV-GENOME-0377588_188 | 98.100 | 1.127E-100 | 317 | 1.000% | MGV | PF08010 | phrog_ | ||
| IMGVR_UViG_2706796236_000001|2706796236|2708709336 | 96.000 | 4.536E-98 | 309 | 1.000% | IMGVR | PF08010 | phrog_ | ||
| OQ174502_00046 | 98.000 | 1.545E-100 | 316 | 1.000% | Escherichia phage REP3 | Escherichia | INPHARED | PF08010 | phrog_ |
| OP172786_00217 | 98.200 | 8.219E-101 | 317 | 1.000% | Escherichia phage ECO07P3 | Escherichia | INPHARED | PF08010 | phrog_ |
| OK272473_00220 | 96.900 | 3.630E-99 | 312 | 1.000% | Escherichia phage JLBYU31 | Escherichia | INPHARED | PF08010 | phrog_ |
| MT446392_00080 | 87.600 | 1.632E-87 | 279 | 1.000% | Escherichia virus TH15 | Escherichia | INPHARED | PF08010 | phrog_ |
| MK373784_00211 | 95.300 | 4.133E-97 | 306 | 1.000% | Escherichia phage vB_EcoM_MM02 | Escherichia | INPHARED | PF08010 | phrog_ |
| LN881728_00075 | 97.700 | 3.984E-100 | 315 | 1.000% | Escherichia phage slur03 | Escherichia | INPHARED | PF08010 | phrog_ |
| GQ981382_00216 | 88.100 | 3.368E-88 | 281 | 1.000% | Shigella phage SP18 | Shigella | INPHARED | PF08010 | phrog_ |
| MGV-GENOME-0377591_192 | 97.900 | 2.119E-100 | 316 | 1.000% | MGV | PF08010 | phrog_ | ||
| OP172785_00116 | 96.500 | 1.283E-98 | 311 | 1.000% | Escherichia phage ECO07P2 | Escherichia | INPHARED | PF08010 | phrog_ |
| OM960734_00091 | 94.600 | 2.747E-96 | 304 | 1.000% | Escherichia phage vB_EcoM_SCS57 | Escherichia | INPHARED | PF08010 | phrog_652 |
| MK373781_00207 | 97.700 | 3.984E-100 | 315 | 1.000% | Escherichia phage vB_EcoM_KAW1E185 | Escherichia | INPHARED | PF08010 | phrog_ |
| KM501444_00214 | 96.400 | 1.759E-98 | 310 | 1.000% | Shigella phage pSs-1 | Shigella | INPHARED | PF08010 | phrog_ |
| IMGVR_UViG_3300045988_132814|3300045988|Ga0495776_001669_29523_29903 | 96.300 | 4.129E-80 | 258 | 0.829% | IMGVR | PF08010 | phrog_ | ||
| OQ599309_00209 | 98.000 | 1.545E-100 | 316 | 1.000% | Escherichia phage 107 | Escherichia | INPHARED | PF08010 | phrog_ |
| MW882907_00198 | 95.600 | 1.603E-97 | 308 | 1.000% | Escherichia phage vB_EcoM_SQ17 | Escherichia | INPHARED | PF08010 | phrog_ |
| MK373779_00207 | 98.100 | 1.127E-100 | 317 | 1.000% | Escherichia phage vB_EcoM_G9062 | Escherichia | INPHARED | PF08010 | phrog_ |
| MH791409_00025 | 86.400 | 5.258E-86 | 274 | 1.000% | Escherichia phage EcWhh-1 | Escherichia | INPHARED | PF08010 | phrog_ |
| OQ031495_00061 | 96.400 | 1.759E-98 | 310 | 1.000% | Escherichia phage MRostami-2023e | Escherichia | INPHARED | PF08010 | phrog_ |
| ON528744_00224 | 97.800 | 2.905E-100 | 316 | 1.000% | Shigella phage ESh34 | Shigella | INPHARED | PF08010 | phrog_ |
| OL964745_00202 | 96.400 | 1.759E-98 | 310 | 1.000% | Escherichia phage T4 | Escherichia | INPHARED | PF08010 | phrog_ |
| OL956808_00038 | 97.800 | 2.905E-100 | 316 | 1.000% | Escherichia phage vB_EcoM-S1P5QW | Escherichia | INPHARED | PF08010 | phrog_ |
| MK373775_00196 | 94.800 | 1.461E-96 | 305 | 1.000% | Escherichia phage vB_EcoM_WFK | Escherichia | INPHARED | PF08010 | phrog_ |
| MF001359_00205 | 97.000 | 2.647E-99 | 313 | 1.000% | Escherichia phage EC121 | Escherichia | INPHARED | PF08010 | phrog_ |
| OM632671_00279 | 95.300 | 4.133E-97 | 306 | 1.000% | Escherichia phage BEU50 | Escherichia | INPHARED | PF08010 | phrog_652 |
| ON528743_00138 | 98.300 | 5.994E-101 | 317 | 1.000% | Shigella phage ESh33 | Shigella | INPHARED | PF08010 | phrog_ |
| MT446387_00047 | 96.000 | 4.536E-98 | 309 | 1.000% | Escherichia phage TH09 | Escherichia | INPHARED | PF08010 | phrog_ |
| KX828711_00201 | 97.500 | 7.490E-100 | 314 | 1.000% | Shigella phage SH7 | Shigella | INPHARED | PF08010 | phrog_ |
| IMGVR_UViG_648936714_000001|648936714|648937009 | 89.400 | 7.625E-90 | 286 | 1.000% | IMGVR | PF08010 | phrog_ | ||
| ON528742_00194 | 96.700 | 6.825E-99 | 312 | 1.000% | Shigella phage ESh32 | Shigella | INPHARED | PF08010 | phrog_ |
| OL964735_00204 | 98.100 | 1.127E-100 | 317 | 1.000% | Escherichia phage T4 | Escherichia | INPHARED | PF08010 | phrog_ |
| MW822007_00191 | 94.900 | 1.066E-96 | 305 | 1.000% | Escherichia phage BF15 | Escherichia | INPHARED | PF08010 | phrog_ |
| uvig_249051_4 | 97.900 | 2.119E-100 | 316 | 1.000% | GPD | PF08010 | phrog_ | ||
| uvig_374229_232 | 94.400 | 5.165E-96 | 303 | 1.000% | GPD | PF08010 | phrog_ | ||
| OQ446694_00231 | 96.000 | 4.536E-98 | 309 | 1.000% | Escherichia phage Killian | Escherichia | INPHARED | PF08010 | phrog_ |
| OQ174503_00022 | 97.900 | 2.119E-100 | 316 | 1.000% | Escherichia phage REP4 | Escherichia | INPHARED | PF08010 | phrog_ |
| ON528741_00047 | 97.500 | 7.490E-100 | 314 | 1.000% | Shigella phage ESh31 | Shigella | INPHARED | PF08010 | phrog_ |
| OV877511_00028 | 95.800 | 8.528E-98 | 308 | 1.000% | Escherichia phage UGJNEcP2 | Escherichia | INPHARED | PF08010 | phrog_ |
| MZ501106_00038 | 97.800 | 2.905E-100 | 316 | 1.000% | Escherichia phage TadeuszReichstein | Escherichia | INPHARED | PF08010 | phrog_ |
| DQ904452_00201 | 97.900 | 2.119E-100 | 316 | 1.000% | Escherichia phage RB32 | Escherichia | INPHARED | PF08010 | phrog_ |
| MK234886_00156 | 86.900 | 9.010E-85 | 271 | 0.980% | Escherichia phage AnYang | Escherichia | INPHARED | PF08010 | phrog_ |
| OQ174500_00042 | 98.400 | 4.372E-101 | 318 | 1.000% | Escherichia phage REP1 | Escherichia | INPHARED | PF08010 | phrog_ |
| KX009778_00204 | 98.400 | 4.372E-101 | 318 | 1.000% | Escherichia phage UFV-AREG1 | Escherichia | INPHARED | PF08010 | phrog_ |
| ON782582_00147 | 95.300 | 4.133E-97 | 306 | 1.000% | Escherichia phage ECP52 | Escherichia | INPHARED | PF08010 | phrog_ |
| ON528739_00128 | 97.100 | 1.931E-99 | 313 | 1.000% | Shigella phage ESh29 | Shigella | INPHARED | PF08010 | phrog_ |
| OM386655_00190 | 88.500 | 1.306E-88 | 282 | 1.000% | Escherichia phage vB_EcoM_ESCO10 | Escherichia | INPHARED | PF08010 | phrog_ |
| OV877085_00268 | 97.300 | 1.027E-99 | 314 | 1.000% | Escherichia phage UGKSEcP1 | Escherichia | INPHARED | PF08010 | phrog_ |
| KM606999_00204 | 98.000 | 1.545E-100 | 316 | 1.000% | Enterobacteria phage RB10 | Enterobacteria | INPHARED | PF08010 | phrog_ |
| MGV-GENOME-0377341_80 | 97.700 | 3.984E-100 | 315 | 1.000% | MGV | PF08010 | phrog_ | ||
| ON528738_00060 | 97.100 | 1.931E-99 | 313 | 1.000% | Shigella phage ESh28 | Shigella | INPHARED | PF08010 | phrog_ |
| OV876900_00220 | 96.900 | 3.630E-99 | 312 | 1.000% | Escherichia phage UGKSEcP2 | Escherichia | INPHARED | PF08010 | phrog_ |
| MW749000_00201 | 97.300 | 1.027E-99 | 314 | 1.000% | Hafnia phage vB_HpaM_IsaacDaniel | Hafnia | INPHARED | PF08010 | phrog_ |
| MW822011_00093 | 96.000 | 4.536E-98 | 309 | 1.000% | Shigella phage TB004 | Shigella | INPHARED | PF08010 | phrog_ |
| OP611477_00137 | 97.300 | 1.027E-99 | 314 | 1.000% | Escherichia virus CAM-21 | Escherichia | INPHARED | PF08010 | phrog_ |
| MZ501096_00037 | 97.700 | 3.984E-100 | 315 | 1.000% | Escherichia phage Paracelsus | Escherichia | INPHARED | PF08010 | phrog_652 |
| MGV-GENOME-0371733_189 | 98.500 | 3.188E-101 | 318 | 1.000% | MGV | PF08010 | phrog_ | ||
| OK331996_00096 | 97.100 | 1.931E-99 | 313 | 1.000% | Escherichia phage vB_EcoM-G157lw | Escherichia | INPHARED | PF08010 | phrog_ |
| ON528735_00194 | 96.600 | 9.358E-99 | 311 | 1.000% | Shigella phage ESh25 | Shigella | INPHARED | PF08010 | phrog_ |
| MT630408_00207 | 96.700 | 1.602E-80 | 259 | 0.829% | Enterobacteria phage phiC600P9 | Enterobacteria | INPHARED | PF08010 | phrog_ |
| OL615012_00204 | 98.000 | 1.545E-100 | 316 | 1.000% | Shigella phage vB_SboM_Phaginator | Shigella | INPHARED | PF08010 | phrog_ |
| OQ067376_00037 | 97.100 | 6.213E-81 | 260 | 0.829% | Escherichia phage a51 | Escherichia | INPHARED | PF08010 | phrog_ |
| OP870146_00207 | 96.100 | 3.308E-98 | 310 | 1.000% | Escherichia phage vB_Eco_F27 | Escherichia | INPHARED | PF08010 | phrog_ |
| OP555982_00206 | 95.400 | 3.014E-97 | 307 | 1.000% | Escherichia phage vB_EcoM-UFV11 | Escherichia | INPHARED | PF08010 | phrog_ |
| MN850609_00057 | 86.800 | 1.488E-86 | 276 | 1.000% | Escherichia phage dhaeg | Escherichia | INPHARED | PF08010 | phrog_ |
| IMGVR_UViG_2974739471_000001|2974739471|2974739687 | 96.000 | 4.536E-98 | 309 | 1.000% | IMGVR | PF08010 | phrog_ | ||
| IMGVR_UViG_651703089_000001|651703089|651709887 | 96.400 | 3.012E-80 | 258 | 0.829% | IMGVR | PF08010 | phrog_ | ||
| OQ067375_00037 | 98.000 | 1.545E-100 | 316 | 1.000% | Escherichia phage a48 | Escherichia | INPHARED | PF08010 | phrog_652 |
| OP870145_00208 | 98.200 | 8.219E-101 | 317 | 1.000% | Escherichia phage vB_Eco_F26 | Escherichia | INPHARED | PF08010 | phrog_ |
| ON528734_00265 | 97.900 | 2.119E-100 | 316 | 1.000% | Shigella phage ESh24 | Shigella | INPHARED | PF08010 | phrog_ |
| uvig_174994_27 | 88.500 | 1.306E-88 | 282 | 1.000% | GPD | PF08010 | phrog_ | ||
| IMGVR_UViG_2595698700_000001|2595698700|2596507262 | 96.800 | 1.168E-80 | 259 | 0.829% | IMGVR | PF08010 | phrog_ | ||
| IMGVR_UViG_2974741780_000001|2974741780|2974741985 | 96.800 | 1.168E-80 | 259 | 0.829% | IMGVR | PF08010 | phrog_ | ||
| OQ067374_00039 | 95.700 | 1.169E-97 | 308 | 1.000% | Escherichia phage a20 | Escherichia | INPHARED | PF08010 | phrog_652 |
| OP870144_00204 | 98.200 | 8.219E-101 | 317 | 1.000% | Escherichia phage vB_Eco_F25 | Escherichia | INPHARED | PF08010 | phrog_ |
| MT515756_00037 | 97.000 | 2.647E-99 | 313 | 1.000% | Yersinia phage PYps55T | Yersinia | INPHARED | PF08010 | phrog_ |
| MK524178_00027 | 96.100 | 3.308E-98 | 310 | 1.000% | Escherichia phage PHB12 | Escherichia | INPHARED | PF08010 | phrog_ |
| AP018932_00205 | 97.200 | 1.408E-99 | 314 | 1.000% | Escherichia phage KIT03 | Escherichia | INPHARED | PF08010 | phrog_ |
| OQ067373_00037 | 97.200 | 4.531E-81 | 260 | 0.829% | Escherichia phage a15 | Escherichia | INPHARED | PF08010 | phrog_ |
| MZ417521_00205 | 97.700 | 3.984E-100 | 315 | 1.000% | Escherichia phage 132 | Escherichia | INPHARED | PF08010 | phrog_ |
| MT968995_00197 | 95.800 | 8.528E-98 | 308 | 1.000% | Escherichia phage vB_EcoM_WL-3 | Escherichia | INPHARED | PF08010 | phrog_ |
| MT611523_00050 | 88.000 | 4.618E-88 | 280 | 1.000% | Escherichia phage DK-13 | Escherichia | INPHARED | PF08010 | phrog_ |
| MH992510_00207 | 96.700 | 1.602E-80 | 259 | 0.829% | Escherichia phage vB_EcoM_DalCa | Escherichia | INPHARED | PF08010 | phrog_ |
| KX130865_00122 | 97.300 | 1.027E-99 | 314 | 1.000% | Shigella phage SHSML-52-1 | Shigella | INPHARED | PF08010 | phrog_ |
| KP007362_00215 | 87.900 | 6.332E-88 | 280 | 1.000% | Escherichia phage vB_EcoM_VR26 | Escherichia | INPHARED | PF08010 | phrog_ |
| OQ223305_00161 | 97.900 | 2.119E-100 | 316 | 1.000% | Escherichia phage EO1 | Escherichia | INPHARED | PF08010 | phrog_ |
| MW598459_00239 | 88.300 | 1.791E-88 | 282 | 1.000% | Escherichia phage vB_EcoM_RZ | Escherichia | INPHARED | PF08010 | phrog_ |
| OK335775_00110 | 88.100 | 3.368E-88 | 281 | 1.000% | Escherichia phage PSD2002 | Escherichia | INPHARED | PF08010 | phrog_ |
| MZ189262_00203 | 97.900 | 2.119E-100 | 316 | 1.000% | Escherichia phage W143 | Escherichia | INPHARED | PF08010 | phrog_ |
| HM071924_00213 | 89.400 | 7.625E-90 | 286 | 1.000% | Escherichia phage IME08 | Escherichia | INPHARED | PF08010 | phrog_ |
| AdfN | 100.000 | 3.188E-101 | 318 | 1.000% | Escherichia phage T4 | Escherichia | NCBI | PF08010 | phrog_ |
| OL770074_00186 | 95.500 | 2.503E-95 | 301 | 0.980% | Escherichia phage vB_EcoM-RPN187 | Escherichia | INPHARED | PF08010 | phrog_ |
| MZ501067_00038 | 98.100 | 1.127E-100 | 317 | 1.000% | Escherichia phage FriedrichZschokke | Escherichia | INPHARED | PF08010 | phrog_ |
| MZ189261_00216 | 97.000 | 2.647E-99 | 313 | 1.000% | Escherichia phage S143_2 | Escherichia | INPHARED | PF08010 | phrog_ |
| LR746311_00212 | 97.800 | 2.905E-100 | 316 | 1.000% | Shigella phage vB_SsoM_113 | Shigella | INPHARED | PF08010 | phrog_ |
| MN716856_00014 | 96.800 | 1.168E-80 | 259 | 0.829% | Yersinia phage vB_YepM_ZN18 | Yersinia | INPHARED | PF08010 | phrog_ |
| MK047717_00037 | 95.800 | 8.528E-98 | 308 | 1.000% | Escherichia phage p000v | Escherichia | INPHARED | PF08010 | phrog_ |
| MH051917_00074 | 85.200 | 1.694E-84 | 270 | 1.000% | Enterobacteria phage vB_EcoM_IME341 | Enterobacteria | INPHARED | PF08010 | phrog_ |
| JN202312_00202 | 97.500 | 7.490E-100 | 314 | 1.000% | Escherichia phage ime09 | Escherichia | INPHARED | PF08010 | phrog_ |
| OQ721911_00208 | 97.300 | 1.027E-99 | 314 | 1.000% | Escherichia phage vB_VIPECOOM03 | Escherichia | INPHARED | PF08010 | phrog_ |
| OP292222_00029 | 86.500 | 3.835E-86 | 275 | 1.000% | Escherichia phage Esco_phi_B-01 | Escherichia | INPHARED | PF08010 | phrog_ |
| OL362041_00209 | 97.100 | 1.931E-99 | 313 | 1.000% | Escherichia phage vB_Eco_OMNI12 | Escherichia | INPHARED | PF08010 | phrog_ |
| MZ501065_00038 | 98.100 | 1.127E-100 | 317 | 1.000% | Escherichia phage FelixPlatter | Escherichia | INPHARED | PF08010 | phrog_ |
| MW269952_00201 | 88.100 | 3.368E-88 | 281 | 1.000% | Escherichia phage P479 | Escherichia | INPHARED | PF08010 | phrog_ |
| MK639187_00251 | 96.800 | 4.977E-99 | 312 | 1.000% | Shigella phage SSE1 | Shigella | INPHARED | PF08010 | phrog_ |
| MH051916_00070 | 96.400 | 1.759E-98 | 310 | 1.000% | Enterobacteria phage vB_EcoM_IME340 | Enterobacteria | INPHARED | PF08010 | phrog_ |
| KF208315_00203 | 98.100 | 1.127E-100 | 317 | 1.000% | Yersinia phage PST | Yersinia | INPHARED | PF08010 | phrog_ |
| OL656107_00208 | 97.200 | 1.408E-99 | 314 | 1.000% | Salmonella phage UAB_60 | Salmonella | INPHARED | PF08010 | phrog_ |
| MT129654_00256 | 86.400 | 5.258E-86 | 274 | 1.000% | Escherichia phage MN04 | Escherichia | INPHARED | PF08010 | phrog_ |
| MH051915_00072 | 96.700 | 6.825E-99 | 312 | 1.000% | Enterobacteria phage vB_EcoM_IME339 | Enterobacteria | INPHARED | PF08010 | phrog_ |
| ON720975_00263 | 95.800 | 8.528E-98 | 308 | 1.000% | Salmonella phage GRNsp7 | Salmonella | INPHARED | PF08010 | phrog_ |
| ON528728_00183 | 97.800 | 2.905E-100 | 316 | 1.000% | Shigella phage ESh18 | Shigella | INPHARED | PF08010 | phrog_ |
| MW679410_00214 | 88.000 | 4.618E-88 | 280 | 1.000% | Escherichia phage C6 | Escherichia | INPHARED | PF08010 | phrog_ |
| MT129652_00291 | 87.300 | 4.208E-87 | 278 | 1.000% | Escherichia phage MN01 | Escherichia | INPHARED | PF08010 | phrog_ |
| MT176425_00209 | 98.200 | 8.219E-101 | 317 | 1.000% | Citrobacter phage PhiZZ23 | Citrobacter | INPHARED | PF08010 | phrog_ |
| KX452698_00164 | 96.600 | 9.358E-99 | 311 | 1.000% | Shigella phage KNP5 | Shigella | INPHARED | PF08010 | phrog_ |
| uvig_280983_244 | 97.800 | 2.905E-100 | 316 | 1.000% | GPD | PF08010 | phrog_ | ||
| OQ466432_00188 | 97.800 | 2.905E-100 | 316 | 1.000% | Escherichia phage ADUt | Escherichia | INPHARED | PF08010 | phrog_652 |
| MZ501056_00037 | 96.800 | 4.977E-99 | 312 | 1.000% | Escherichia phage ChristianSchoenbein | Escherichia | INPHARED | PF08010 | phrog_ |
| MK327942_00205 | 96.800 | 4.977E-99 | 312 | 1.000% | Escherichia phage vB_EcoM_G50 | Escherichia | INPHARED | PF08010 | phrog_ |
| KU925172_00063 | 97.500 | 7.490E-100 | 314 | 1.000% | Escherichia phage PE37 | Escherichia | INPHARED | PF08010 | phrog_ |
| MGV-GENOME-0377517_152 | 94.400 | 5.165E-96 | 303 | 1.000% | MGV | PF08010 | phrog_ | ||
| IMGVR_UViG_3300045988_090047|3300045988|Ga0495776_101649_65187_65567 | 96.100 | 5.662E-80 | 257 | 0.829% | IMGVR | PF08010 | phrog_ | ||
| IMGVR_UViG_2765235844_000001|2765235844|2765591635 | 88.100 | 3.368E-88 | 281 | 1.000% | IMGVR | PF08010 | phrog_ | ||
| OL770107_00232 | 86.900 | 1.085E-86 | 276 | 1.000% | Escherichia phage CLB_P2 | Escherichia | INPHARED | PF08010 | phrog_ |
| MZ726795_00206 | 97.900 | 2.119E-100 | 316 | 1.000% | Escherichia phage vB_EcoM_ASO78A | Escherichia | INPHARED | PF08010 | phrog_ |
| OK274152_00205 | 97.600 | 5.462E-100 | 315 | 1.000% | Escherichia phage APTC-EC-2A | Escherichia | INPHARED | PF08010 | phrog_ |
| MZ501047_00037 | 96.100 | 3.308E-98 | 310 | 1.000% | Escherichia phage AlbertHofmann | Escherichia | INPHARED | PF08010 | phrog_ |
| LC620505_00005 | 96.600 | 9.358E-99 | 311 | 1.000% | Escherichia phage NiC89 | Escherichia | INPHARED | PF08010 | phrog_ |
| MW233709_00205 | 88.300 | 1.791E-88 | 282 | 1.000% | Escherichia phage UPEC01 | Escherichia | INPHARED | PF08010 | phrog_ |
| KU878969_00203 | 87.300 | 4.208E-87 | 278 | 1.000% | Escherichia phage MX01 | Escherichia | INPHARED | PF08010 | phrog_ |
| OP856590_00203 | 97.700 | 3.984E-100 | 315 | 1.000% | Escherichia phage Ec_MI-02 | Escherichia | INPHARED | PF08010 | phrog_ |
| MK327934_00209 | 96.200 | 2.412E-98 | 310 | 1.000% | Escherichia phage vB_EcoM_G2469 | Escherichia | INPHARED | PF08010 | phrog_ |
| MT682713_00206 | 97.600 | 5.462E-100 | 315 | 1.000% | Escherichia phage vB_EcoM_Ozark | Escherichia | INPHARED | PF08010 | phrog_ |
| MK327933_00203 | 97.200 | 1.408E-99 | 314 | 1.000% | Escherichia phage vB_EcoM_G2285 | Escherichia | INPHARED | PF08010 | phrog_ |
| IMGVR_UViG_2974734346_000001|2974734346|2974734355 | 96.900 | 8.520E-81 | 259 | 0.829% | IMGVR | PF08010 | phrog_ | ||
| MZ150757_00208 | 98.000 | 1.545E-100 | 316 | 1.000% | Salmonella phage Lv5cm | Salmonella | INPHARED | PF08010 | phrog_ |
| MT682710_00097 | 97.900 | 2.119E-100 | 316 | 1.000% | Escherichia phage vB_EcoM_FT | Escherichia | INPHARED | PF08010 | phrog_ |
| MN781580_00206 | 97.600 | 5.462E-100 | 315 | 1.000% | Shigella phage KRT47 | Shigella | INPHARED | PF08010 | phrog_ |
| MK962755_00206 | 97.500 | 7.490E-100 | 314 | 1.000% | Shigella phage JK38 | Shigella | INPHARED | PF08010 | phrog_ |
| MK327929_00206 | 97.200 | 1.408E-99 | 314 | 1.000% | Escherichia phage D5505 | Escherichia | INPHARED | PF08010 | phrog_ |
| KR233165_00211 | 96.100 | 3.308E-98 | 310 | 1.000% | Escherichia phage PEC04 | Escherichia | INPHARED | PF08010 | phrog_ |