

| Basic Information | |
|---|---|
| Species | Fragaria vesca |
| Cazyme ID | mrna24180.1-v1.0-hybrid |
| Family | GH35 |
| Protein Properties | Length: 670 Molecular Weight: 76123.1 Isoelectric Point: 6.7165 |
| Chromosome | Chromosome/Scaffold: 1 Start: 10698713 End: 10703358 |
| Description | Glycosyl hydrolase family 35 protein |
| View CDS | |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH35 | 37 | 338 | 0 |
| LIINGQRRILFSGSIHYPRSTPEMWPSLIGKAKEGGLDVIQTYVFWNLHEPQPGQYDFSGRYDLVKFIKEIQAQGLYACLRIGPFIESEWTYGGFPFWLH DVPGIVYRTDNEPFKNFTTKIVNLMKSEGLYASQGGPIILSQVENEYQNVEAAFHEKGPAYVRWAAKMAVDLQTGVPWVMCKQTDAPDPVVNTCNGMQCG RTFGGPNSPNKPSMWTENWTSFYQVYGGEPYIRSAEDIAFHVALFIARMNGSYVNYYMYHGGTNFGRTSSAYVITGYYDQAPLDEYGLLRQPKWGHLKQL HA | |||
| Full Sequence |
|---|
| Protein Sequence Length: 670 Download |
| MERQRWLRFW VMVVVVLITD EGAFVTGGNV TYDGRSLIIN GQRRILFSGS IHYPRSTPEM 60 WPSLIGKAKE GGLDVIQTYV FWNLHEPQPG QYDFSGRYDL VKFIKEIQAQ GLYACLRIGP 120 FIESEWTYGG FPFWLHDVPG IVYRTDNEPF KNFTTKIVNL MKSEGLYASQ GGPIILSQVE 180 NEYQNVEAAF HEKGPAYVRW AAKMAVDLQT GVPWVMCKQT DAPDPVVNTC NGMQCGRTFG 240 GPNSPNKPSM WTENWTSFYQ VYGGEPYIRS AEDIAFHVAL FIARMNGSYV NYYMYHGGTN 300 FGRTSSAYVI TGYYDQAPLD EYGLLRQPKW GHLKQLHAAV RNCSTTLLQG VRTNISLGQL 360 QEAYVFQEES AGRCVAFLIN NDEKVNATVQ FRNISYYLRP KSISILPDCG KVIFNTAKVN 420 TSYNKRITRT VQIFDSVDRW EGSKDIIQNF EPTISLQSDT LLEHMNVTKD KSDYLWYTMR 480 FQHNISCTEP ILHVESLGHV THAFVNGIYT GNAHGSHDTK QFIMENPIVL NNGSNNISLL 540 SVMVGLPDSG AFLESKFAGL RRVEIHCNET DQFYNFTSYA WGYQVGLLGQ KLQIYREENL 600 DDFEWSNIEG SFNKPLTWYK TYFDAPMGND PVTFNLSSMA KGEAWINGQS IGRYWVSFLT 660 SKGKPSQTM* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
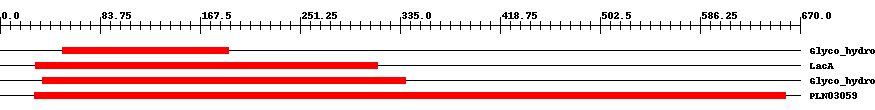 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam02449 | Glyco_hydro_42 | 2.0e-6 | 52 | 191 | 158 | + Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. | ||
| COG1874 | LacA | 9.0e-19 | 30 | 316 | 330 | + Beta-galactosidase [Carbohydrate transport and metabolism] | ||
| pfam01301 | Glyco_hydro_35 | 3.0e-152 | 36 | 340 | 323 | + Glycosyl hydrolases family 35. | ||
| PLN03059 | PLN03059 | 0 | 29 | 658 | 647 | + beta-galactosidase; Provisional | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds |
| GO:0004565 | beta-galactosidase activity |
| GO:0005975 | carbohydrate metabolic process |
| GO:0009341 | beta-galactosidase complex |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3d3a_A | 9e-37 | 21 | 336 | 5 | 325 | A Chain A, Crystal Structure Of A Beta-Galactosidase From Bacteroides Thetaiotaomicron |
| PDB | 3thd_D | 4e-30 | 30 | 327 | 11 | 322 | A Chain A, Crystal Structure Of A Beta-Galactosidase From Bacteroides Thetaiotaomicron |
| PDB | 3thd_C | 4e-30 | 30 | 327 | 11 | 322 | A Chain A, Crystal Structure Of A Beta-Galactosidase From Bacteroides Thetaiotaomicron |
| PDB | 3thd_B | 4e-30 | 30 | 327 | 11 | 322 | A Chain A, Crystal Structure Of A Beta-Galactosidase From Bacteroides Thetaiotaomicron |