
| APIS family ID | APIS107 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inhibited defense system | restriction-modification (RM) | |||||||||||||
| CLAN ID | CLAN045 | |||||||||||||
| Seed protein property |
|
|||||||||||||
| Phage property |
|
|||||||||||||
| PMID/References | PMID:6307815 | |||||||||||||
| PDB structures | ; | |||||||||||||
| Pfam domains | PF02086.20,PF02086.20 | |||||||||||||
| Phrog | ||||||||||||||
| Host taxa | d__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacterales;f__Enterobacteriaceae;g__Escherichia; | |||||||||||||
| Gene Location | Start: 16846; End: 17625; Strand: - | |||||||||||||
| Description | DNA adenine methylases methylate adenine residues in specific sequences, the methylation site of the host Escherichia coli dam+ methylase. Methylation protects the site against cleavage by the MboI restriction nuclease. | |||||||||||||
| Genome accession | Start | End | Strand | Protein ID | Length | Molecular weight | Charge | Isoelectric point | Function | Phrog | PFAM |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NC_000866.4 | 14685 | 15026 | - | NP_049642.1 | 113 | 12620.65 | -10.5 | 3.6524 | Mrh.1 hypothetical protein | phrog_2031 | |
| NC_000866.4 | 15026 | 15232 | - | NP_049643.1 | 68 | 8257.51 | -1.0 | 5.6578 | Mrh.2 hypothetical protein | phrog_1873 | |
| NC_000866.4 | 15331 | 15573 | - | NP_049644.1 | 80 | 9117.17 | 0.0 | 6.4636 | virion structural protein | phrog_1715 | PF16855.10 |
| NC_000866.4 | 15606 | 16280 | - | NP_049645.1 | 224 | 26216.13 | 30.5 | 10.7389 | SegF homing endonuclease | ||
| NC_000866.4 | 16270 | 16785 | - | NP_049646.1 | 171 | 20424.18 | -10.5 | 4.5572 | dUTPase | PF08761.16 | |
| NC_000866.4 | 16846 | 17625 | - | NP_049647.1 | 259 | 30416.80 | 9.5 | 9.2073 | Dam family site-specific DNA-(adenine-N6)-methyltransferase | PF02086.20 ;PF02086.20 | |
| NC_000866.4 | 17935 | 18963 | - | NP_049648.1 | 342 | 39777.99 | 15.5 | 9.5801 | DNA primase | phrog_21887 | PF08275.16 |
| NC_000866.4 | 18966 | 19130 | - | NP_049649.1 | 54 | 5896.64 | -1.0 | 5.0824 | gp61.1 hypothetical protein | phrog_2086 ; phrog_1823 | |
| NC_000866.4 | 19132 | 19758 | - | NP_049650.1 | 208 | 24331.57 | 0.0 | 6.4890 | gp61.2 hypothetical protein | phrog_4122 ; phrog_3781 ; phrog_8539 | |
| NC_000866.4 | 19758 | 20051 | - | NP_049651.1 | 97 | 10993.66 | -3.0 | 4.6017 | spackle periplasmic | phrog_1586 | |
| NC_000866.4 | 20112 | 20369 | - | NP_049652.1 | 85 | 10187.10 | 8.5 | 10.4193 | gp61.4 hypothetical protein | phrog_12748 ; phrog_3272 ; phrog_2375 ; phrog_8921 |
pTM is for the estimate of the TM-score, which is obtained from a pairwise error prediction. The higher pTM score indicates better model quality.
pLDDT is for per-residue accuracy of the structure, which representes the quality of the residue. A higher value indicates better prediction accuracy. More detail please see AlphaFold .
| Hit | Query start | Query end | Hit start | Hit end | TMscore(ali) | TMscore(avg) |
|---|---|---|---|---|---|---|
| AF-Q8ZMS3-F1-model_v4 | 1 | 259 | 1 | 260 | 0.8634 | 0.8139 |
| AF-Q32AJ9-F1-model_v4 | 1 | 259 | 3 | 268 | 0.8522 | 0.8112 |
| AF-P0AEE8-F1-model_v4 | 1 | 259 | 3 | 268 | 0.8518 | 0.8107 |
| AF-A0A0H3GWR3-F1-model_v4 | 1 | 259 | 3 | 270 | 0.847 | 0.8106 |
| AF-P0DMP3-F1-model_v4 | 1 | 259 | 3 | 268 | 0.8509 | 0.8101 |
| AF-Q58015-F1-model_v4 | 1 | 259 | 2 | 288 | 0.8488 | 0.7901 |
| AF-P44243-F1-model_v4 | 1 | 259 | 23 | 296 | 0.7003 | 0.6346 |
| AF-Q5F5M8-F1-model_v4 | 1 | 259 | 4 | 332 | 0.7305 | 0.6318 |
| Hit | Query start | Query end | Hit start | Hit end | TMscore(ali) | TMscore(avg) |
|---|---|---|---|---|---|---|
| MGYP000259042174.pdb | 1 | 259 | 4 | 279 | 0.9301 | 0.8877 |
| MGYP000271406668.pdb | 1 | 259 | 3 | 279 | 0.9299 | 0.8861 |
| MGYP000994564167.pdb | 1 | 259 | 2 | 278 | 0.9274 | 0.886 |
| MGYP001000926617.pdb | 1 | 259 | 5 | 270 | 0.914 | 0.8851 |
| MGYP003571243029.pdb | 1 | 259 | 2 | 284 | 0.9349 | 0.8848 |
| MGYP001024232661.pdb | 5 | 259 | 1 | 273 | 0.9182 | 0.8845 |
| MGYP000860657186.pdb | 1 | 259 | 3 | 282 | 0.9294 | 0.8817 |
| MGYP000249947192.pdb | 1 | 259 | 3 | 262 | 0.8967 | 0.8811 |
| MGYP000042165803.pdb | 1 | 239 | 2 | 260 | 0.8881 | 0.8785 |
| MGYP000897117520.pdb | 9 | 259 | 1 | 263 | 0.8971 | 0.8784 |
| Fam ID | Seed protein | Member count | Model | Alignment | APIS107 | NP_049647.1 | 110 | HMM model | Member alignment |
|---|
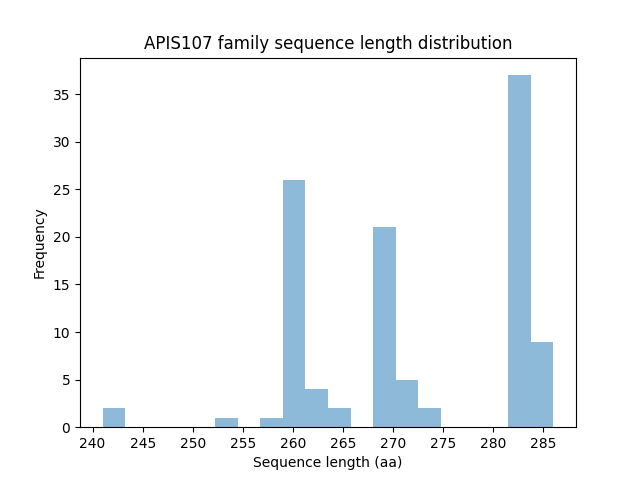
| Member | pident | evalue | bitscore | qcovs | phage | host | SourceDB | Pfam | Phrog |
|---|---|---|---|---|---|---|---|---|---|
| MN871507_00003 | 61.400 | 1.907E-97 | 314 | 1.000% | Aeromonas phage asfd_1h | Aeromonas | INPHARED | PF02086 | phrog_ |
| ON042478_00093 | 61.400 | 2.612E-97 | 313 | 1.000% | Aeromonas phage AhFM11 | Aeromonas | INPHARED | PF02086 | phrog_ |
| MGV-GENOME-0377561_28 | 63.500 | 8.037E-102 | 326 | 1.000% | MGV | PF02086 | phrog_ | ||
| MGV-GENOME-0377499_93 | 62.900 | 1.367E-100 | 323 | 1.000% | MGV | PF02086 | phrog_ | ||
| OK499996_00268 | 62.600 | 6.597E-100 | 321 | 1.000% | Klebsiella phage vB_KaeM_Shinkou | Klebsiella | INPHARED | PF02086 | phrog_ |
| LR877331_00120 | 62.600 | 4.815E-100 | 321 | 1.000% | Klebsiella phage vB_KpnM_311F | Klebsiella | INPHARED | PF02086 | phrog_ |
| KY000080_00021 | 63.500 | 8.037E-102 | 326 | 1.000% | Klebsiella phage KPV15 | Klebsiella | INPHARED | PF02086 | phrog_ |
| OM337876_00278 | 63.500 | 8.037E-102 | 326 | 1.000% | Klebsiella phage KP182 | Klebsiella | INPHARED | PF02086 | phrog_ |
| IMGVR_UViG_3300045988_090046|3300045988|Ga0495776_001918_66348_67133 | 62.900 | 1.367E-100 | 323 | 1.000% | IMGVR | PF02086 | phrog_ | ||
| MH729874_00251 | 63.400 | 1.101E-101 | 326 | 1.000% | Klebsiella phage KP179 | Klebsiella | INPHARED | PF02086 | phrog_ |
| MN038175_00040 | 49.100 | 2.101E-73 | 244 | 0.996% | Panteoa phage Phynn | Pantoea | INPHARED | PF02086 | phrog_ |
| MF327006_00203 | 43.800 | 7.351E-61 | 208 | 0.996% | Shigella phage Sf20 | Shigella | INPHARED | PF02086 | phrog_27806 |
| MK962753_00065 | 44.200 | 1.120E-61 | 210 | 0.996% | Shigella phage JK32 | Shigella | INPHARED | PF02086 | phrog_27806 |
| MW748999_00192 | 44.200 | 8.181E-62 | 211 | 0.996% | Escherichia phage vB_EcoM_Skers | Escherichia | INPHARED | PF02086 | phrog_27806 |
| MN709127_00061 | 44.100 | 1.532E-61 | 210 | 0.996% | Escherichia phage Ec_Makalu_002 | Escherichia | INPHARED | PF02086 | phrog_27806 |
| OX001802_00201 | 44.500 | 2.333E-62 | 212 | 0.996% | Escherichia phage vB_Eco_TB34 | Escherichia | INPHARED | PF02086 | phrog_27806 |
| MGV-GENOME-0375882_202 | 44.200 | 1.120E-61 | 210 | 0.996% | MGV | PF02086 | phrog_27806 | ||
| ON286974_00061 | 44.200 | 1.120E-61 | 210 | 0.996% | Escherichia phage W115 | Escherichia | INPHARED | PF02086 | phrog_27806 |
| MT764208_00182 | 44.000 | 2.869E-61 | 209 | 0.996% | Escherichia phage JEP8 | Escherichia | INPHARED | PF02086 | phrog_ |
| MK573636_00071 | 43.300 | 6.603E-60 | 205 | 0.996% | Escherichia phage vB_EcoM_PHB13 | Escherichia | INPHARED | PF02086 | phrog_27806 |
| MN850615_00204 | 44.200 | 8.181E-62 | 211 | 0.996% | Escherichia phage kvi | Escherichia | INPHARED | PF02086 | phrog_27806 |
| HE978309_00063 | 43.700 | 1.006E-60 | 207 | 0.996% | Enterobacteria phage GEC-3S | Enterobacteria | INPHARED | PF02086 | phrog_ |
| uvig_374832_55 | 44.500 | 2.333E-62 | 212 | 0.996% | GPD | PF02086 | phrog_27806 | ||
| MH560358_00221 | 44.700 | 6.650E-63 | 214 | 0.996% | Escherichia virus KFS-EC | Escherichia | INPHARED | PF02086 | phrog_ |
| MN894885_00061 | 44.200 | 1.120E-61 | 210 | 0.996% | Escherichia phage Ec_Makalu_001 | Escherichia | INPHARED | PF02086 | phrog_27806 |
| uvig_100047_108 | 44.200 | 1.120E-61 | 210 | 0.996% | GPD | PF02086 | phrog_27806 | ||
| MN882349_00061 | 44.300 | 3.192E-62 | 212 | 1.000% | Escherichia phage Ec_Makalu_003 | Escherichia | INPHARED | PF02086 | phrog_27806 |
| uvig_589117_33 | 44.000 | 2.097E-61 | 209 | 0.996% | GPD | PF02086 | phrog_ | ||
| MZ504878_00062 | 44.300 | 5.978E-62 | 211 | 0.996% | Enterobacteria phage Whisky | Enterobacteria | INPHARED | PF02086 | phrog_ |
| MZ504877_00064 | 44.500 | 2.333E-62 | 212 | 0.996% | Enterobacteria phage Cognac | Enterobacteria | INPHARED | PF02086 | phrog_ |
| MZ504876_00060 | 44.300 | 5.978E-62 | 211 | 0.996% | Enterobacteria phage Brandy | Enterobacteria | INPHARED | PF02086 | phrog_27806 |
| OP114734_00205 | 43.800 | 7.351E-61 | 208 | 0.996% | Escherichia phage vB_EcoM-pEE20 | Escherichia | INPHARED | PF02086 | phrog_ |
| KY683735_00065 | 44.700 | 6.650E-63 | 214 | 0.996% | Escherichia phage ECD7 | Escherichia | INPHARED | PF02086 | phrog_ |
| MT478991_00246 | 44.300 | 5.978E-62 | 211 | 0.996% | Escherichia phage vB_EcoM_011D4 | Escherichia | INPHARED | PF02086 | phrog_27806 |
| MK327947_00065 | 44.000 | 2.097E-61 | 209 | 0.996% | Escherichia phage vB_EcoM_G5211 | Escherichia | INPHARED | PF02086 | phrog_ |
| MK327935_00064 | 44.200 | 1.120E-61 | 210 | 0.996% | Escherichia phage vB_EcoM_G2494 | Escherichia | INPHARED | PF02086 | phrog_27806 |
| AY343333_00062 | 44.100 | 1.532E-61 | 210 | 0.996% | Escherichia phage RB49 | Escherichia | INPHARED | PF02086 | phrog_27806 |
| MGV-GENOME-0374910_27 | 54.000 | 2.808E-82 | 270 | 0.992% | MGV | PF02086 | phrog_ | ||
| OP684128_00242 | 54.000 | 2.808E-82 | 270 | 0.992% | Klebsiella phage pR7_1 | Klebsiella | INPHARED | PF02086 | phrog_ |
| MW021756_00241 | 52.900 | 3.140E-80 | 264 | 0.996% | Cronobacter phage vB_CsaM_SemperBestia | Cronobacter | INPHARED | PF02086 | phrog_ |
| OL551674_00260 | 53.300 | 3.476E-81 | 267 | 0.996% | Enterobacter phage vB_EcRAM-01 | Enterobacter | INPHARED | PF02086 | phrog_ |
| MW021749_00167 | 51.400 | 4.338E-77 | 255 | 0.996% | Citrobacter phage vB_Cfr_Xman | Citrobacter | INPHARED | PF02086 | phrog_ |
| OQ571799_00156 | 51.700 | 9.009E-78 | 257 | 0.996% | Enterobacter phage phi5 | Enterobacter | INPHARED | PF02086 | phrog_ |
| OX335407_00186 | 54.100 | 1.497E-82 | 271 | 0.992% | Klebsiella phage mtp7 | Klebsiella | INPHARED | PF02086 | phrog_ |
| JN882287_00242 | 53.300 | 3.476E-81 | 267 | 0.996% | Cronobacter phage vB_CsaM_GAP161 | Cronobacter | INPHARED | PF02086 | phrog_ |
| MZ612130_00055 | 54.100 | 2.051E-82 | 270 | 0.992% | Klebsiella phage vB_KpnM-VAC66 | Klebsiella | INPHARED | PF02086 | phrog_ |
| MH823906_00246 | 53.100 | 1.223E-80 | 265 | 0.996% | Citrobacter phage Maroon | Citrobacter | INPHARED | PF02086 | phrog_ |
| KT321315_00245 | 53.600 | 1.853E-81 | 267 | 0.992% | Enterobacter phage phiEap-3 | Enterobacter | INPHARED | PF02086 | phrog_ |
| MN508624_00245 | 53.700 | 5.268E-82 | 269 | 0.996% | Enterobacter phage EC-F2 | Enterobacter | INPHARED | PF02086 | phrog_ |
| MN101224_00030 | 54.100 | 2.051E-82 | 270 | 0.992% | Klebsiella phage KMI10 | Klebsiella | INPHARED | PF02086 | phrog_ |
| HE858210_00249 | 52.200 | 9.977E-79 | 259 | 0.996% | Pseudotevenvirus RB43 | Unspecified | INPHARED | PF02086 | phrog_ |
| uvig_581689_78 | 53.300 | 3.476E-81 | 267 | 0.996% | GPD | PF02086 | phrog_ | ||
| OP970827_00082 | 53.300 | 4.760E-81 | 266 | 0.996% | Klebsiella phage Kp_GWPR59 | Klebsiella | INPHARED | PF02086 | phrog_ |
| OP045497_00016 | 53.700 | 1.353E-81 | 268 | 0.992% | Klebsiella phage 150040 | Klebsiella | INPHARED | PF02086 | phrog_ |
| MN508622_00233 | 52.800 | 4.301E-80 | 263 | 0.996% | Enterobacter phage EC-W2 | Enterobacter | INPHARED | PF02086 | phrog_ |
| OK210075_00075 | 53.600 | 7.214E-82 | 269 | 0.996% | Citrobacter phage KKP_3664 | Citrobacter | INPHARED | PF02086 | phrog_ |
| IMGVR_UViG_2974557979_000001|2974557979|2974558229 | 52.000 | 3.206E-65 | 220 | 0.857% | Enterobacteriaceae | IMGVR | PF02086 | phrog_ | |
| HM134276_00240 | 53.300 | 3.476E-81 | 267 | 0.996% | Escherichia phage RB16 | Escherichia | INPHARED | PF02086 | phrog_ |
| IMGVR_UViG_2974587579_000001|2974587579|2974587826 | 53.300 | 2.340E-71 | 238 | 0.900% | IMGVR | PF02086 | phrog_ | ||
| IMGVR_UViG_648936715_000001|648936715|648937927 | 51.200 | 7.395E-64 | 216 | 0.857% | IMGVR | PF02086 | phrog_ | ||
| MW250785_00039 | 53.100 | 1.223E-80 | 265 | 0.996% | Escherichia phage UPEC03 | Escherichia | INPHARED | PF02086 | phrog_ |
| HQ918180_00241 | 53.700 | 9.880E-82 | 268 | 0.992% | Klebsiella phage KP27 | Klebsiella | INPHARED | PF02086 | phrog_ |
| AY967407_00253 | 51.600 | 1.689E-77 | 256 | 0.996% | Escherichia phage RB43 | Escherichia | INPHARED | PF02086 | phrog_ |
| MGV-GENOME-0378091_26 | 54.000 | 2.808E-82 | 270 | 0.992% | MGV | PF02086 | phrog_ | ||
| MGV-GENOME-0375107_27 | 53.300 | 3.476E-81 | 267 | 0.996% | MGV | PF02086 | phrog_ | ||
| MGV-GENOME-0378171_221 | 54.200 | 1.093E-82 | 271 | 0.992% | MGV | PF02086 | phrog_ | ||
| KC801932_00237 | 52.300 | 7.285E-79 | 260 | 0.996% | Escherichia phage Lw1 | Escherichia | INPHARED | PF02086 | phrog_ |
| OP617746_00184 | 54.000 | 2.808E-82 | 270 | 0.992% | Klebsiella phage KP13MC5-1 | Klebsiella | INPHARED | PF02086 | phrog_ |
| KX245890_00240 | 51.100 | 2.089E-76 | 253 | 0.996% | Citrobacter phage vB_CfrM_CfP1 | Citrobacter | INPHARED | PF02086 | phrog_ |
| ON157416_00242 | 53.100 | 1.223E-80 | 265 | 0.996% | Cronobacter phage Dev_CS701 | Cronobacter | INPHARED | PF02086 | phrog_ |
| OL539470_00021 | 52.800 | 4.301E-80 | 263 | 0.996% | Cronobacter phage vB_CsaM_Invicta | Cronobacter | INPHARED | PF02086 | phrog_ |
| MN508817_00198 | 50.100 | 1.242E-74 | 248 | 1.000% | Yersinia phage JC221 | Yersinia | INPHARED | PF02086 | phrog_ |
| KX443552_00251 | 53.400 | 2.538E-81 | 267 | 0.996% | Cronobacter phage vB_CsaM_leE | Cronobacter | INPHARED | PF02086 | phrog_ |
| KR869820_00232 | 51.500 | 3.168E-77 | 255 | 0.996% | Citrobacter phage IME-CF2 | Citrobacter | INPHARED | PF02086 | phrog_ |
| IMGVR_UViG_3300015157_000003|3300015157|Ga0181864_12499820 | 54.200 | 1.093E-82 | 271 | 0.992% | IMGVR | PF02086 | phrog_ | ||
| LR746310_00257 | 53.800 | 7.214E-82 | 269 | 0.992% | Klebsiella phage vB_KpnM_05F | Klebsiella | INPHARED | PF02086 | phrog_ |
| MW448170_00246 | 54.100 | 1.497E-82 | 271 | 0.992% | Klebsiella phage vB_KpnM_M1 | Klebsiella | INPHARED | PF02086 | phrog_ |
| KX431559_00246 | 53.500 | 1.353E-81 | 268 | 0.996% | Cronobacter phage vB_CsaM_leB | Cronobacter | INPHARED | PF02086 | phrog_ |
| IMGVR_UViG_3300045988_041714|3300045988|Ga0495776_110494_15088_15939 | 54.000 | 2.808E-82 | 270 | 0.992% | IMGVR | PF02086 | phrog_ | ||
| MT334653_00251 | 52.300 | 7.285E-79 | 260 | 0.996% | Buttiauxella phage vB_ButM_GuL6 | Buttiauxella | INPHARED | PF02086 | phrog_ |
| OQ267591_00256 | 53.700 | 1.353E-81 | 268 | 0.992% | Klebsiella phage phi_KPN_S3 | Klebsiella | INPHARED | PF02086 | phrog_ |
| ON515458_00244 | 54.200 | 1.093E-82 | 271 | 0.992% | Klebsiella phage BL02 | Klebsiella | INPHARED | PF02086 | phrog_ |
| MZ322895_00079 | 53.800 | 7.214E-82 | 269 | 0.992% | Klebsiella phage vB_KpnM_VAC13 | Klebsiella | INPHARED | PF02086 | phrog_ |
| KY971610_00204 | 52.900 | 3.508E-78 | 258 | 0.992% | Pseudomonas phage PspYZU05 | Pseudomonas | INPHARED | PF02086 | phrog_ |
| MGV-GENOME-0377602_140 | 95.700 | 4.115E-170 | 523 | 1.000% | MGV | PF02086 | phrog_ | ||
| KR269718_00173 | 96.300 | 2.410E-171 | 527 | 1.000% | Escherichia phage HY03 | Escherichia | INPHARED | PF02086 | phrog_ |
| OM135583_00028 | 95.700 | 4.115E-170 | 523 | 1.000% | Escherichia phage EP01 | Escherichia | INPHARED | PF02086 | phrog_ |
| MK373785_00030 | 95.700 | 4.115E-170 | 523 | 1.000% | Escherichia phage vB_EcoM_OE5505 | Escherichia | INPHARED | PF02086 | phrog_ |
| OQ599309_00030 | 96.000 | 8.506E-171 | 525 | 1.000% | Escherichia phage 107 | Escherichia | INPHARED | PF02086 | phrog_ |
| NP_049647.1 | 100.000 | 9.098E-175 | 537 | 1.000% | Escherichia phage T4 | Escherichia | NCBI | PF02086 PF02086 | phrog_ |
| OL964735_00031 | 96.900 | 1.030E-172 | 531 | 1.000% | Escherichia phage T4 | Escherichia | INPHARED | PF02086 | phrog_ |
| MG867727_00030 | 96.000 | 1.166E-170 | 525 | 1.000% | Escherichia phage vB_EcoM-G28 | Escherichia | INPHARED | PF02086 | phrog_ |
| MF001354_00203 | 96.300 | 2.410E-171 | 527 | 1.000% | Salmonella phage SG1 | Salmonella | INPHARED | PF02086 | phrog_ |
| uvig_585812_256 | 95.800 | 3.002E-170 | 524 | 1.000% | GPD | PF02086 | phrog_ | ||
| MW822011_00276 | 95.600 | 7.730E-170 | 522 | 1.000% | Shigella phage TB004 | Shigella | INPHARED | PF02086 | phrog_ |
| ON528734_00172 | 96.000 | 1.166E-170 | 525 | 1.000% | Shigella phage ESh24 | Shigella | INPHARED | PF02086 | phrog_ |
| MZ501089_00141 | 95.500 | 1.060E-169 | 522 | 1.000% | Escherichia phage KarlGJung | Escherichia | INPHARED | PF02086 | phrog_ |
| ON229909_00235 | 96.100 | 4.528E-171 | 526 | 1.000% | Escherichia phage vB_EcoM_CE1 | Escherichia | INPHARED | PF02086 | phrog_ |
| KX130861_00215 | 96.100 | 4.528E-171 | 526 | 1.000% | Shigella phage SHFML-11 | Shigella | INPHARED | PF02086 | phrog_ |
| OL362041_00032 | 95.600 | 5.640E-170 | 523 | 1.000% | Escherichia phage vB_Eco_OMNI12 | Escherichia | INPHARED | PF02086 | phrog_ |
| IMGVR_UViG_3300006879_000208|3300006879|Ga0079226_100114192 | 95.900 | 1.598E-170 | 524 | 1.000% | IMGVR | PF02086 | phrog_ | ||
| ON528727_00058 | 94.700 | 4.658E-168 | 517 | 1.000% | Shigella phage ESh17 | Shigella | INPHARED | PF02086 | phrog_ |
| KU878969_00027 | 75.600 | 2.889E-127 | 400 | 1.000% | Escherichia phage MX01 | Escherichia | INPHARED | PF02086 | phrog_ |
| KU925172_00237 | 95.100 | 7.025E-169 | 520 | 1.000% | Escherichia phage PE37 | Escherichia | INPHARED | PF02086 | phrog_ |
| IMGVR_UViG_3300019879_000001|3300019879|Ga0193723_100006840 | 43.200 | 1.006E-60 | 207 | 0.996% | IMGVR | PF02086 | phrog_ | ||
| IMGVR_UViG_3300031418_000373|3300031418|Ga0308166_10137639 | 43.100 | 2.577E-60 | 206 | 0.996% | IMGVR | PF02086 | phrog_ | ||
| IMGVR_UViG_3300045988_067560|3300045988|Ga0495776_014908_94011_94832 | 54.700 | 7.983E-83 | 271 | 1.000% | IMGVR | PF02086 | phrog_ | ||
| MGV-GENOME-0378626_206 | 55.000 | 2.269E-83 | 273 | 1.000% | MGV | PF02086 | phrog_ | ||
| OM638103_00097 | 55.000 | 2.269E-83 | 273 | 1.000% | Cronobacter phage LPCS28 | Cronobacter | INPHARED | PF02086 | phrog_ |
| MT560058_00035 | 52.200 | 4.804E-78 | 258 | 1.000% | Pectobacterium phage POP12 | Pectobacterium | INPHARED | PF02086 | phrog_ |