

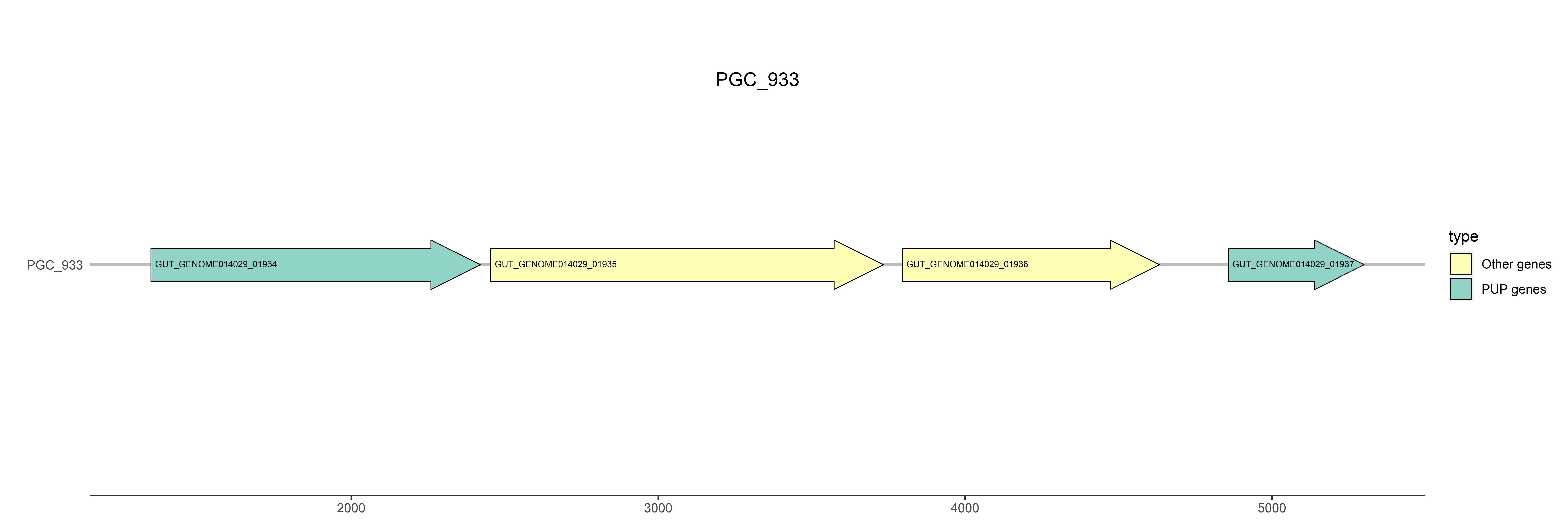
Gene: iolG_1
Representative genome: GUT_GENOME014029
Gene type: PUP genes
Position: 1347..2420
Protein length: 358
Product: Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase
EC: 1.1.1.18
Pfam: Glyco_hydro_3, Glyco_hydro_3_C, Fn3-like, Gly_kinase, TetR_N
Lineage: Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Agathobacter;
Geography: Fiji, Oceania
Gene: iolG_2
Representative genome: GUT_GENOME014029
Gene type: other genes
Position: 2454..3734
Protein length: 427
Product: Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase
EC: 1.1.1.18
Pfam: Glyco_hydro_3, Glyco_hydro_3_C, Fn3-like, Gly_kinase, TetR_N
Lineage: Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Agathobacter;
Geography: Fiji, Oceania
Gene: iolE
Representative genome: GUT_GENOME014029
Gene type: other genes
Position: 3795..4634
Protein length: 280
Product: Inosose dehydratase
EC: 4.2.1.44
Pfam: Glyco_hydro_106, Glyco_hydro_2_N
Lineage: Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Agathobacter;
Geography: Fiji, Oceania
Gene: NA
Representative genome: GUT_GENOME014029
Gene type: PUP genes
Position: 4857..5300
Protein length: 148
Product: hypothetical protein
EC: NA
Pfam: Glyco_hydro_106, Glyco_hydro_2_N
Lineage: Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Agathobacter;
Geography: Fiji, Oceania