

| Title | H2O2 Production in Species of the Lactobacillus acidophilus Group: a Central Role for a Novel NADH-Dependent Flavin Reductase |
|---|---|
| Author | Rosanne Hertzberger, Jos Arents, Henk L. Dekker, R. David Pridmore, Christof Gysler, Michiel Kleerebezem, M. Joost Teixeira de Mattos |
| DOI | 10.1128/AEM.04272-13 |
| Abstract | Hydrogen peroxide production is a well-known trait of many bacterial species associated with the human body. In the presence of oxygen, the probiotic lactic acid bacterium Lactobacillus johnsonii NCC 533 excretes up to 1 mM H2O2, inducing growth stagnation and cell death. Disruption of genes commonly assumed to be involved in H2O2 production (e.g., pyruvate oxidase, NADH oxidase, and lactate oxidase) did not affect this. Here we describe the purification of a novel NADH-dependent flavin reductase encoded by two highly similar genes (LJ0548 and LJ0549) that are conserved in lactobacilli belonging to the Lactobacillus acidophilus group. The genes are predicted to encode two 20-kDa proteins containing flavin mononucleotide (FMN) reductase conserved domains. Reductase activity requires FMN, flavin adenine dinucleotide (FAD), or riboflavin and is specific for NADH and not NADPH. The Km for FMN is 30 ± 8 μM, in accordance with its proposed in vivo role in H2O2 production. Deletion of the encoding genes in L. johnsonii led to a 40-fold reduction of hydrogen peroxide formation. H2O2 production in this mutant could only be restored by in trans complementation of both genes. Our work identifies a novel, conserved NADH-dependent flavin reductase that is prominently involved in H2O2 production in L. johnsonii. |
Uniprot ID: Q74HL7
Protein: NADH-dependent flavin reductase subunit 1
Organism: Lactobacillus johnsonii (strain CNCM I-12250 / La1 / NCC 533)
Length: 178 AA
Taxonomic identifier: 257314 [NCBI]
| Source | Domain | Start | End | E-value (Domain) | Coverage |
|---|---|---|---|---|---|
| Pfam-A | FMN_red | 1 | 144 | 2.1e-29 | 0.981 |
Program: hmmscan
Version: 3.1b2 (February 2015)
Method: hmmscan --domtblout hmmscan.tbl --noali -E 1e-5 pfam query.fa
Date: Mon Jul 20 14:32:16 2020
Description:
FMN_red
No Pfam abstract.
This domain in found in several flavoproteins such as FMN-dependent NADPH-azoreductase, which catalyses the reductive cleavage of azo bond in aromatic azo compounds to the corresponding amines[1], and NAD§H:quinone oxidoreductase, which reduces quinones to the hydroquinone state to prevent interaction of the semiquinone with O2 and production of superoxide[2]. In Arabidopsis NADPH:quinone oxidoreductase is involved in detoxification pathways[3]. NAD§H:quinone oxidoreductase prefers NADH over NADPH, while FMN-dependent NADPH-azoreductase requires NADPH, but not NADH, as an electron donor for its activity.
Other proteins with this domain include iron-sulfur flavoproteins[4] and chromate reductase[5].
Information is taken from Pfam and InterPro web site.
flavin mononucleotide + NADH + H+ ⇒ 1,5-Dihydroriboflavin 5’-(dihydrogen phosphate) + NAD+
![[L1]flavin mononucleotide](../static/images/chemical_structure/OR7/[L1]flavin mononucleotide.png)

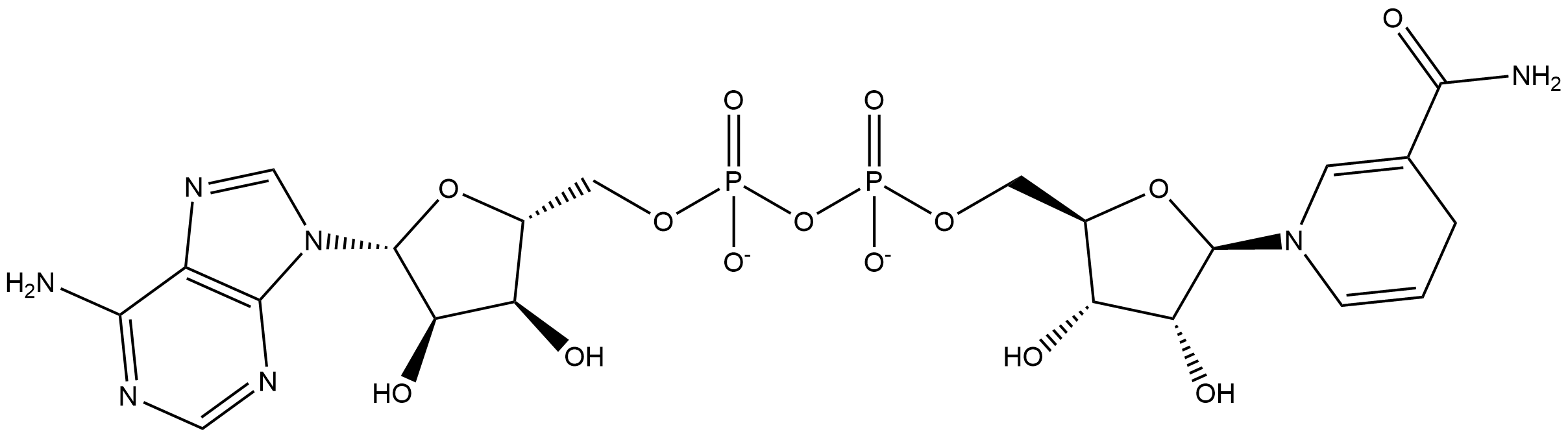

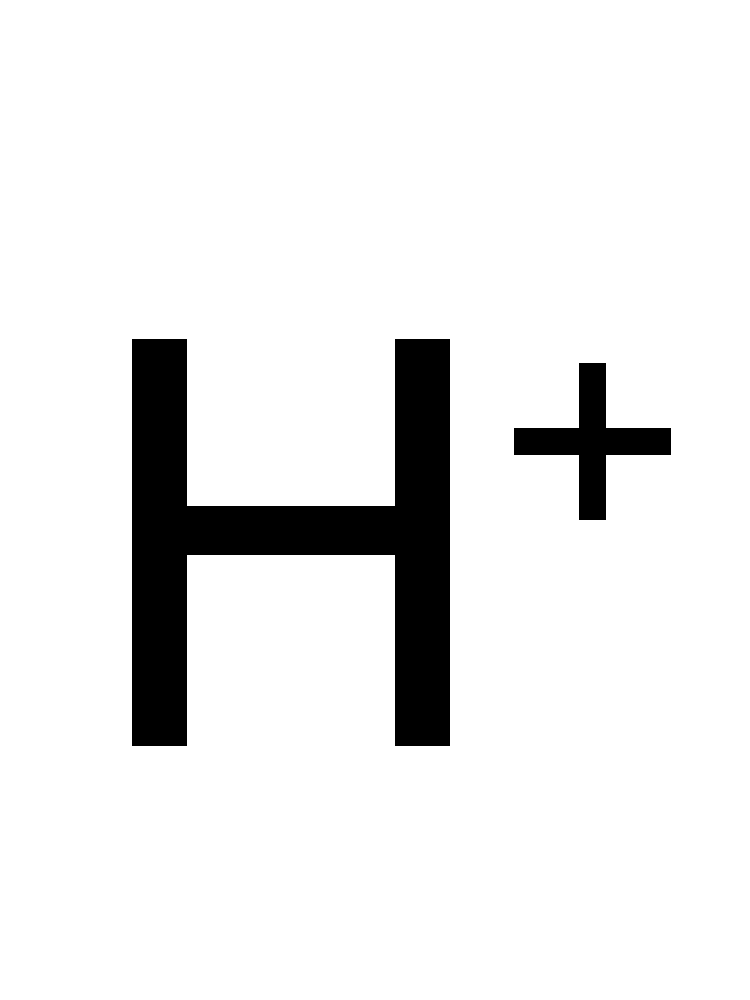

![[R1]1,5-Dihydroriboflavin 5'-(dihydrogen phosphate)](../static/images/chemical_structure/OR7/[R1]1,5-Dihydroriboflavin 5'-(dihydrogen phosphate).png)

![[R2]NAD+](../static/images/chemical_structure/OR7/[R2]NAD+.png)
Deller S, Sollner S, Trenker-El-Toukhy R, et al. Characterization of a thermostable NADPH: FMN oxidoreductase from the mesophilic bacterium Bacillus subtilis[J]. Biochemistry, 2006, 45(23): 7083-7091. ↩︎
Grandori R, Khalifah P, Boice J A, et al. Biochemical characterization of WrbA, founding member of a new family of multimeric flavodoxin-like proteins[J]. Journal of Biological Chemistry, 1998, 273(33): 20960-20966. ↩︎
Sparla F, Tedeschi G, Pupillo P, et al. Cloning and heterologous expression of NAD § H: quinone reductase of Arabidopsis thaliana, a functional homologue of animal DT-diaphorase[J]. FEBS letters, 1999, 463(3): 382-386. ↩︎
Zhao T, Cruz F, Ferry J G. Iron-Sulfur Flavoprotein (Isf) fromMethanosarcina thermophila Is the Prototype of a Widely Distributed Family[J]. Journal of Bacteriology, 2001, 183(21): 6225-6233. ↩︎
Ackerley D F, Gonzalez C F, Park C H, et al. Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli[J]. Applied and environmental microbiology, 2004, 70(2): 873-882. ↩︎
| Title | H2O2 Production in Species of the Lactobacillus acidophilus Group: a Central Role for a Novel NADH-Dependent Flavin Reductase |
|---|---|
| Author | Rosanne Hertzberger, Jos Arents, Henk L. Dekker, R. David Pridmore, Christof Gysler, Michiel Kleerebezem, M. Joost Teixeira de Mattos |
| DOI | 10.1128/AEM.04272-13 |
| Abstract | Hydrogen peroxide production is a well-known trait of many bacterial species associated with the human body. In the presence of oxygen, the probiotic lactic acid bacterium Lactobacillus johnsonii NCC 533 excretes up to 1 mM H2O2, inducing growth stagnation and cell death. Disruption of genes commonly assumed to be involved in H2O2 production (e.g., pyruvate oxidase, NADH oxidase, and lactate oxidase) did not affect this. Here we describe the purification of a novel NADH-dependent flavin reductase encoded by two highly similar genes (LJ0548 and LJ0549) that are conserved in lactobacilli belonging to the Lactobacillus acidophilus group. The genes are predicted to encode two 20-kDa proteins containing flavin mononucleotide (FMN) reductase conserved domains. Reductase activity requires FMN, flavin adenine dinucleotide (FAD), or riboflavin and is specific for NADH and not NADPH. The Km for FMN is 30 ± 8 μM, in accordance with its proposed in vivo role in H2O2 production. Deletion of the encoding genes in L. johnsonii led to a 40-fold reduction of hydrogen peroxide formation. H2O2 production in this mutant could only be restored by in trans complementation of both genes. Our work identifies a novel, conserved NADH-dependent flavin reductase that is prominently involved in H2O2 production in L. johnsonii. |
Uniprot ID: Q74HL8
Protein: NADH-dependent flavin reductase subunit 2
Organism: Lactobacillus johnsonii (strain CNCM I-12250 / La1 / NCC 533)
Length: 184 AA
Taxonomic identifier: 257314 [NCBI]
| Source | Domain | Start | End | E-value (Domain) | Coverage |
|---|---|---|---|---|---|
| Pfam-A | FMN_red | 1 | 149 | 7e-30 | 0.987 |
Program: hmmscan
Version: 3.1b2 (February 2015)
Method: hmmscan --domtblout hmmscan.tbl --noali -E 1e-5 pfam query.fa
Date: Mon Jul 20 14:32:16 2020
Description:
FMN_red
No Pfam abstract.
This domain in found in several flavoproteins such as FMN-dependent NADPH-azoreductase, which catalyses the reductive cleavage of azo bond in aromatic azo compounds to the corresponding amines[1], and NAD§H:quinone oxidoreductase, which reduces quinones to the hydroquinone state to prevent interaction of the semiquinone with O2 and production of superoxide[2]. In Arabidopsis NADPH:quinone oxidoreductase is involved in detoxification pathways[3]. NAD§H:quinone oxidoreductase prefers NADH over NADPH, while FMN-dependent NADPH-azoreductase requires NADPH, but not NADH, as an electron donor for its activity.
Other proteins with this domain include iron-sulfur flavoproteins[4] and chromate reductase[5].
Information is taken from Pfam and InterPro web site.
flavin mononucleotide + NADH + H+ ⇒ 1,5-Dihydroriboflavin 5’-(dihydrogen phosphate) + NAD+
![[L1]flavin mononucleotide](../static/images/chemical_structure/OR7/[L1]flavin mononucleotide.png)



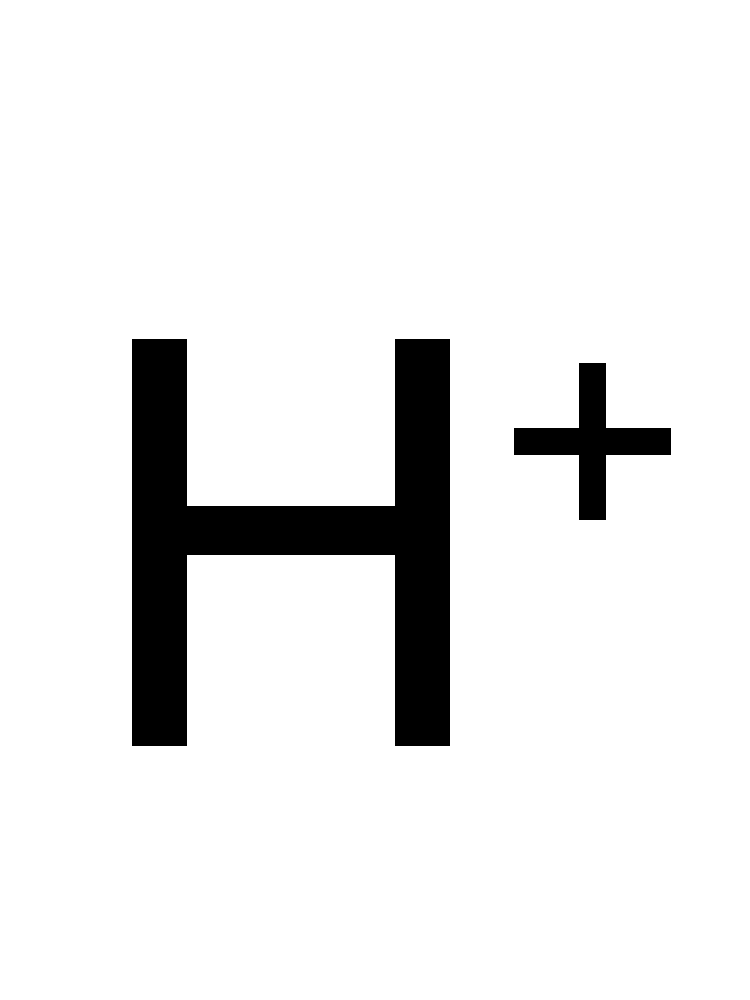

![[R1]1,5-Dihydroriboflavin 5'-(dihydrogen phosphate)](../static/images/chemical_structure/OR7/[R1]1,5-Dihydroriboflavin 5'-(dihydrogen phosphate).png)

![[R2]NAD+](../static/images/chemical_structure/OR7/[R2]NAD+.png)
Deller S, Sollner S, Trenker-El-Toukhy R, et al. Characterization of a thermostable NADPH: FMN oxidoreductase from the mesophilic bacterium Bacillus subtilis[J]. Biochemistry, 2006, 45(23): 7083-7091. ↩︎
Grandori R, Khalifah P, Boice J A, et al. Biochemical characterization of WrbA, founding member of a new family of multimeric flavodoxin-like proteins[J]. Journal of Biological Chemistry, 1998, 273(33): 20960-20966. ↩︎
Sparla F, Tedeschi G, Pupillo P, et al. Cloning and heterologous expression of NAD § H: quinone reductase of Arabidopsis thaliana, a functional homologue of animal DT-diaphorase[J]. FEBS letters, 1999, 463(3): 382-386. ↩︎
Zhao T, Cruz F, Ferry J G. Iron-Sulfur Flavoprotein (Isf) fromMethanosarcina thermophila Is the Prototype of a Widely Distributed Family[J]. Journal of Bacteriology, 2001, 183(21): 6225-6233. ↩︎
Ackerley D F, Gonzalez C F, Park C H, et al. Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli[J]. Applied and environmental microbiology, 2004, 70(2): 873-882. ↩︎