

| Basic Information | |
|---|---|
| Species | Prunus persica |
| Cazyme ID | ppa003656m |
| Family | AA4 |
| Protein Properties | Length: 560 Molecular Weight: 61425.6 Isoelectric Point: 6.4542 |
| Chromosome | Chromosome/Scaffold: 7 Start: 12649970 End: 12658264 |
| Description | FAD-linked oxidases family protein |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| AA4 | 95 | 327 | 1.9e-26 |
| KEVPRELIDELKAICQDNMTLDYEERYNHGKPQNSFHKAVNIPDVVVFPRSEEQVSKIVNSCDKHKVPIVPYGGATSLEGHTLSPNGGVCIDMSLMNRVK ALHVEDMDVVVEPGVGWMELNEYLEPYGLFFPLDPAGPGATIGGMCATRCSGSLAVRYGTMRDNVISLKVVLANGDVVKTASRARKSAAGYDLTRLVIGS EGTLGVITEVTLRLQKIPQYSVVAVCNFPTVKD | |||
| Full Sequence |
|---|
| Protein Sequence Length: 560 Download |
| MAMSSWFSRL RCSSKCFFNS LALRTSVSRK LVRTQTTINS ESTRNPFLFW SRSLLPIALA 60 VSAGSLALHP QSDPSLCEAP SVNSRAELVV KGSHKEVPRE LIDELKAICQ DNMTLDYEER 120 YNHGKPQNSF HKAVNIPDVV VFPRSEEQVS KIVNSCDKHK VPIVPYGGAT SLEGHTLSPN 180 GGVCIDMSLM NRVKALHVED MDVVVEPGVG WMELNEYLEP YGLFFPLDPA GPGATIGGMC 240 ATRCSGSLAV RYGTMRDNVI SLKVVLANGD VVKTASRARK SAAGYDLTRL VIGSEGTLGV 300 ITEVTLRLQK IPQYSVVAVC NFPTVKDAAD VAIATMLSGI QVSRVELLDE VQVRAINIAN 360 GKSLPETPTL MFEFIGTEAY SREQTLIVKK IASEHNGSDF VFAEDPETKK ELWKMRKEAL 420 WACFAMEPNF EAMISDVCVP LSCLAELISR SKQELDASEL ICTVIAHAGD GNFHTVILFD 480 PNNEEHRQEA QRLNHFMVHT ALSMEGTCTG EHGVGTGKMK YLEQELGMEA LKTMKRIKAA 540 LDPNNTMNPG KLIPSHVCF* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PRK11230 | PRK11230 | 4.0e-54 | 136 | 558 | 428 | + glycolate oxidase subunit GlcD; Provisional | ||
| pfam02913 | FAD-oxidase_C | 1.0e-68 | 311 | 552 | 247 | + FAD linked oxidases, C-terminal domain. This domain has a ferredoxin-like fold. | ||
| TIGR00387 | glcD | 2.0e-96 | 140 | 551 | 420 | + glycolate oxidase, subunit GlcD. This protein, the glycolate oxidase GlcD subunit, is similar in sequence to that of several D-lactate dehydrogenases, including that of E. coli. The glycolate oxidase has been found to have some D-lactate dehydrogenase activity [Energy metabolism, Other]. | ||
| COG0277 | GlcD | 3.0e-128 | 104 | 555 | 465 | + FAD/FMN-containing dehydrogenases [Energy production and conversion] | ||
| PLN02805 | PLN02805 | 0 | 9 | 559 | 558 | + D-lactate dehydrogenase [cytochrome] | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0003824 | catalytic activity |
| GO:0008762 | UDP-N-acetylmuramate dehydrogenase activity |
| GO:0016491 | oxidoreductase activity |
| GO:0050660 | flavin adenine dinucleotide binding |
| GO:0055114 | oxidation-reduction process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| RefSeq | NP_001142976.1 | 0 | 86 | 559 | 83 | 555 | hypothetical protein LOC100275430 [Zea mays] |
| RefSeq | NP_568170.1 | 0 | 50 | 559 | 49 | 567 | FAD linked oxidase family protein [Arabidopsis thaliana] |
| RefSeq | XP_002267321.1 | 0 | 1 | 559 | 1 | 566 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002322755.1 | 0 | 87 | 559 | 8 | 480 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002524111.1 | 0 | 1 | 559 | 1 | 555 | d-lactate dehydrogenase, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3pm9_F | 4.00001e-41 | 100 | 553 | 16 | 476 | A Chain A, Crystal Structure Of A Putative Dehydrogenase (Rpa1076) From Rhodopseudomonas Palustris Cga009 At 2.57 A Resolution |
| PDB | 3pm9_E | 4.00001e-41 | 100 | 553 | 16 | 476 | A Chain A, Crystal Structure Of A Putative Dehydrogenase (Rpa1076) From Rhodopseudomonas Palustris Cga009 At 2.57 A Resolution |
| PDB | 3pm9_D | 4.00001e-41 | 100 | 553 | 16 | 476 | A Chain A, Crystal Structure Of A Putative Dehydrogenase (Rpa1076) From Rhodopseudomonas Palustris Cga009 At 2.57 A Resolution |
| PDB | 3pm9_C | 4.00001e-41 | 100 | 553 | 16 | 476 | A Chain A, Crystal Structure Of A Putative Dehydrogenase (Rpa1076) From Rhodopseudomonas Palustris Cga009 At 2.57 A Resolution |
| PDB | 3pm9_B | 4.00001e-41 | 100 | 553 | 16 | 476 | A Chain A, Crystal Structure Of A Putative Dehydrogenase (Rpa1076) From Rhodopseudomonas Palustris Cga009 At 2.57 A Resolution |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
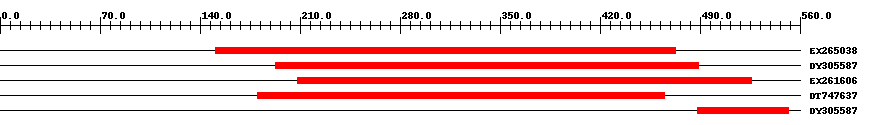 | ||||
| Hit | Length | Start | End | EValue |
| EX265038 | 324 | 151 | 473 | 0 |
| DY305587 | 297 | 193 | 489 | 0 |
| EX261606 | 323 | 208 | 526 | 0 |
| DT747637 | 286 | 180 | 465 | 0 |
| DY305587 | 65 | 488 | 552 | 2.2 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |