

| Basic Information | |
|---|---|
| Species | Prunus persica |
| Cazyme ID | ppa000209m |
| Family | CBM45 |
| Protein Properties | Length: 1468 Molecular Weight: 164234 Isoelectric Point: 6.533 |
| Chromosome | Chromosome/Scaffold: 1 Start: 25148856 End: 25159801 |
| Description | Pyruvate phosphate dikinase, PEP/pyruvate binding domain |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM45 | 462 | 540 | 1.9e-23 |
| LHWALSKNKAGEWSEPPPNALPQGSVSLKGAAETQFQSSADSTYEVQSLEIEIEVESFKGMPFVLCSAGNWIKNQGSDF | |||
| CBM45 | 127 | 202 | 5.2e-25 |
| LHWGGIQDRKEKWVLPSRRPDGTKVYKNKALRTPFQKSGSICLLKIEIDDPAIQAIEFLIVDESQNRWFKNNGDNF | |||
| Full Sequence |
|---|
| Protein Sequence Length: 1468 Download |
| MSNSVGHNLL NQSLLQSKIN SSGIPANTLF QAKSVHQVAA QARKSPISKK FCGNNLNVQK 60 PKSAMGSRHP ATAVPRAVLT TDPPSDLAGK FNLGGNIELQ VYVNASSPGS ATQVEIRVTY 120 SGHSLTLHWG GIQDRKEKWV LPSRRPDGTK VYKNKALRTP FQKSGSICLL KIEIDDPAIQ 180 AIEFLIVDES QNRWFKNNGD NFHVKLPAKE KLISNASVPE ELVQIQAYLR WERKGKQMYT 240 PEQEKVEYEA ARSELLEEVA RGTSIQDLQA RLTKKHDGGK IEEPSLSETK RIPEDLVQIQ 300 SYIRWEKAGK PNYSPEEQHR EFEEARQELQ RELEKGASLD EIRKKITKGE IQTKVAKKFE 360 SKQVFRTDRI QRKKRDFMQI INKQTAKIVD EAKIVDKEHS VKPKPLTAVE LFAKAREEQD 420 GGSVLRKYTF KLNDKDLLVL VTKPAGKTKV HLATDFKEPL TLHWALSKNK AGEWSEPPPN 480 ALPQGSVSLK GAAETQFQSS ADSTYEVQSL EIEIEVESFK GMPFVLCSAG NWIKNQGSDF 540 YVDFGVELKK VQKDAGDGKG TAKGLLDKIA EQESEAQKSF MHRFNIAADL INQATDSGEL 600 GLAGILVWMR FMAMRQLIWN KNYNVKPREI SKAQNRLTDL LQSVYASHPQ YRELLRMIMS 660 TVGRGGEGDV GQRIRDEILV IQRNNECKGG MMEEWHQKLH NNTSPDDVVI CQALLDYIKN 720 DFDIGVYWKT LNDNGITKER LLSYDRAIHN EPNFRRDQKE GLLRDLGHYM RTLKAVHSGA 780 DLESAIQNCM GYKSEGQGFM VGVKINPISG LPSEFPDLLR FVLEHVEDRN VEVLIEGLLE 840 ARQMLWPLLS KPHDRLRDLL FLDIALDSTV RTAIERGYEE LNNAGPEKIM YFISLVLENL 900 ALSSDDNEDL VYCLKGWDHA INMLKSNSDD WALYAKSILD RTRLALANKA ESYLSVLQPS 960 AEYLGSQLGV DQSAVNIFTE EIIRAGSAAS LSSLLNRLDP VLRKTAHLGS WQVISPLEVV 1020 GYVVVVDELL TVQNKVYSKP TILVAKSVKG EEEIPDGTVA VLTPDMPDVL SHVSVRARNS 1080 KVCFATCFDP NILADLQASE GKLLRIKPTP ADITYSEVNE GELEDASSTH STEDIPSLTL 1140 VRKQFTGRYA ISSDEFTSET VGAKSRNIAY IKGKLPSWIG IPTSVALPFG VFEKVLSEDS 1200 NKAVAEKLGT LKKKLKDEDF DSLREIRETV LQLAAPPQLV QELRTKMQSS GMPWPGDEGE 1260 QRWEQAWMAI KKVWASKWNE RAYFSTRKVK LDHDYLCMAV LVQEIINADY AFVIHTTNPS 1320 SGDSSEIYAE VVKGLGETLV GAYPGRALSF ISKKNDLDSP QVLGYPSKPV GLFIRRSIIF 1380 RSDSNGEDLE GYAGAGLYDS VPMDEEEKVV LDYSSDPLMV DGNFRKSILS SIARAGSAIE 1440 ELYGSPQDIE GVIRDGKLYV VQTRPQV* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
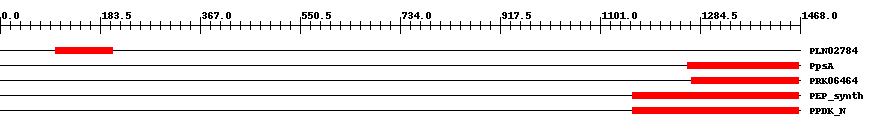 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02784 | PLN02784 | 4.0e-9 | 101 | 207 | 112 | + alpha-amylase | ||
| COG0574 | PpsA | 7.0e-12 | 1262 | 1465 | 208 | + Phosphoenolpyruvate synthase/pyruvate phosphate dikinase [Carbohydrate transport and metabolism] | ||
| PRK06464 | PRK06464 | 3.0e-14 | 1269 | 1465 | 210 | + phosphoenolpyruvate synthase; Validated | ||
| TIGR01418 | PEP_synth | 8.0e-16 | 1161 | 1466 | 343 | + phosphoenolpyruvate synthase. Also called pyruvate,water dikinase and PEP synthase. The member from Methanococcus jannaschii contains a large intein. This enzyme generates phosphoenolpyruvate (PEP) from pyruvate, hydrolyzing ATP to AMP and releasing inorganic phosphate in the process. The enzyme shows extensive homology to other enzymes that use PEP as substrate or product. This enzyme may provide PEP for gluconeogenesis, for PTS-type carbohydrate transport systems, or for other processes [Energy metabolism, Glycolysis/gluconeogenesis]. | ||
| pfam01326 | PPDK_N | 1.0e-32 | 1161 | 1466 | 334 | + Pyruvate phosphate dikinase, PEP/pyruvate binding domain. This enzyme catalyzes the reversible conversion of ATP to AMP, pyrophosphate and phosphoenolpyruvate (PEP). | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005524 | ATP binding |
| GO:0016301 | kinase activity |
| GO:0016310 | phosphorylation |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAK11735.1 | 0 | 1 | 1467 | 1 | 1464 | starch associated protein R1 [Solanum tuberosum] |
| Swiss-Prot | Q8LPT9 | 0 | 1 | 1467 | 1 | 1475 | GWD1_CITRE RecName: Full=Alpha-glucan water dikinase, chloroplastic; AltName: Full=Starch-related protein R1; Flags: Precursor |
| RefSeq | XP_002270485.1 | 0 | 1 | 1467 | 1 | 1470 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002315679.1 | 0 | 2 | 1467 | 5 | 1477 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002527902.1 | 0 | 1 | 1467 | 1 | 1469 | alpha-glucan water dikinase, chloroplast precursor, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2ols_A | 0.0003 | 1159 | 1466 | 18 | 336 | A Chain A, The Crystal Structure Of The Phosphoenolpyruvate Synthase From Neisseria Meningitidis |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
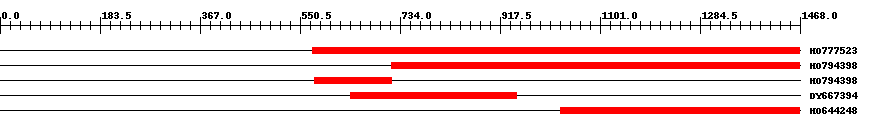 | ||||
| Hit | Length | Start | End | EValue |
| HO777523 | 898 | 573 | 1468 | 0 |
| HO794398 | 752 | 719 | 1468 | 0 |
| HO794398 | 142 | 578 | 719 | 0 |
| DY667394 | 305 | 643 | 947 | 0 |
| HO644248 | 444 | 1028 | 1468 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |