

| Basic Information | |
|---|---|
| Species | Physcomitrella patens |
| Cazyme ID | Pp1s149_288V6.1 |
| Family | CE8 |
| Protein Properties | Length: 573 Molecular Weight: 62634 Isoelectric Point: 8.2164 |
| Chromosome | Chromosome/Scaffold: 149 Start: 1003140 End: 1005425 |
| Description | Plant invertase/pectin methylesterase inhibitor superfamily |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CE8 | 258 | 556 | 0 |
| NVIVAKDGSGKYKTVGEAIQRASTSGATRYVIYVKAGVYDEQIIIPKKLAKLTIIGDGIDKTIFTGKRNVGLMKGMTTYLSATMIVQGEGFIGKMFTCRN TAGAAGHQAVATRVTADKVAFYRVKFDSFQDTLYCHSLRQFYRECIVMGTVDFIFGNANAVFQNCQIVAKKTTLQGQQNTYTAQGRSDKHQNTGLAFQDC NFDGTPDLKRNVQYYPTFLGRPWKAYSVCVLLRPSIQAHVDPKGWLPWNTTDFGLYTSFFAEYKGSGPGSNRRYRVKWSHGISDSKTANKYQAASFIDG | |||
| Full Sequence |
|---|
| Protein Sequence Length: 573 Download |
| MTHDDEKPMV GSYRPKSRRC CVATVLSVSL VVVLVICIAI PLGMRSGKDD GNGDPAACVA 60 TDYPDTCRES LAGSDGTPVG MSKASIKSSD EQVGQLSDGT TNEQCKELLT SARELFARVY 120 ASMDITNQTL REQTCADIQT QLSAALTNVN TCKDVLEESG SPEFASFLER SSKSEQCIGN 180 TLAIVNGFCM YGNEIKNWAG LATGLSMPDL GSFLGNGAHR RLMNVEYSTN EAQEENLLPN 240 WMDSATSRHL LTLPPSYNVI VAKDGSGKYK TVGEAIQRAS TSGATRYVIY VKAGVYDEQI 300 IIPKKLAKLT IIGDGIDKTI FTGKRNVGLM KGMTTYLSAT MIVQGEGFIG KMFTCRNTAG 360 AAGHQAVATR VTADKVAFYR VKFDSFQDTL YCHSLRQFYR ECIVMGTVDF IFGNANAVFQ 420 NCQIVAKKTT LQGQQNTYTA QGRSDKHQNT GLAFQDCNFD GTPDLKRNVQ YYPTFLGRPW 480 KAYSVCVLLR PSIQAHVDPK GWLPWNTTDF GLYTSFFAEY KGSGPGSNRR YRVKWSHGIS 540 DSKTANKYQA ASFIDGKSWI TDLGMPYSNA AV* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02301 | PLN02301 | 2.0e-112 | 239 | 568 | 334 | + pectinesterase/pectinesterase inhibitor | ||
| PLN02745 | PLN02745 | 9.0e-121 | 58 | 566 | 534 | + Putative pectinesterase/pectinesterase inhibitor | ||
| PLN02314 | PLN02314 | 8.0e-122 | 56 | 567 | 541 | + pectinesterase | ||
| PLN02468 | PLN02468 | 4.0e-124 | 46 | 567 | 555 | + putative pectinesterase/pectinesterase inhibitor | ||
| pfam01095 | Pectinesterase | 1.0e-147 | 258 | 556 | 301 | + Pectinesterase. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004857 | enzyme inhibitor activity |
| GO:0005618 | cell wall |
| GO:0030599 | pectinesterase activity |
| GO:0042545 | cell wall modification |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| RefSeq | XP_001758305.1 | 0 | 58 | 566 | 2 | 493 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001767596.1 | 0 | 44 | 570 | 1 | 531 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001767671.1 | 0 | 248 | 570 | 5 | 330 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001772362.1 | 0 | 223 | 572 | 1 | 350 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001783825.1 | 0 | 81 | 568 | 1 | 461 | predicted protein [Physcomitrella patens subsp. patens] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1gq8_A | 0 | 258 | 568 | 8 | 316 | A Chain A, Pectin Methylesterase From Carrot |
| PDB | 1xg2_A | 0 | 258 | 567 | 4 | 311 | B Chain B, Crystal Structure Of The Complex Between Pectin Methylesterase And Its Inhibitor Protein |
| PDB | 1qjv_B | 8e-20 | 256 | 527 | 3 | 311 | A Chain A, Pectin Methylesterase Pema From Erwinia Chrysanthemi |
| PDB | 1qjv_A | 8e-20 | 256 | 527 | 3 | 311 | A Chain A, Pectin Methylesterase Pema From Erwinia Chrysanthemi |
| PDB | 2ntq_B | 8e-20 | 256 | 527 | 3 | 311 | A Chain A, Pectin Methylesterase Pema From Erwinia Chrysanthemi |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| homogalacturonan degradation | RXN-2102 | EC-3.1.1.11 | pectinesterase |
| Transmembrane Domains | ||||
|---|---|---|---|---|
 | ||||
| Start | End | |||
| 21 | 43 | |||
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
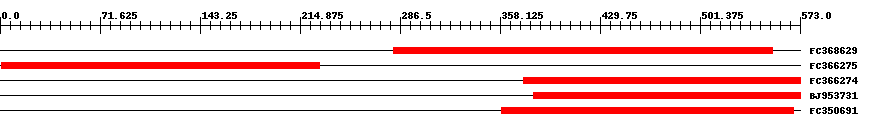 | ||||
| Hit | Length | Start | End | EValue |
| FC368629 | 272 | 282 | 553 | 0 |
| FC366275 | 229 | 1 | 229 | 0 |
| FC366274 | 199 | 375 | 573 | 0 |
| BJ953731 | 192 | 382 | 573 | 0 |
| FC350691 | 210 | 359 | 568 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |