

| Basic Information | |
|---|---|
| Species | Physcomitrella patens |
| Cazyme ID | Pp1s96_59V6.1 |
| Family | CE8 |
| Protein Properties | Length: 602 Molecular Weight: 65621.2 Isoelectric Point: 9.0515 |
| Chromosome | Chromosome/Scaffold: 96 Start: 359662 End: 361859 |
| Description | Plant invertase/pectin methylesterase inhibitor superfamily |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CE8 | 286 | 584 | 0 |
| NVTVAWNGSGKYRKIMDAVKNAPIKSSSPYVIYIKSGIYKEQVKINSSLTNIMLLGDGPAYTIITGSLSVALTKSMTTFLSPTLIVEGQGFKAKGIQVRN TAGPAGHQAVALRVSADKSSFYQCTFDSFQDTLYVHNYRQFYRDCTIKGTIDYIFGNAWAVFQNCRLTAKKSTIVGQVNVYTAQGKTDRGQTTGISFQSC TFDATTDLARNSKAFPTYLGRPWKAYATTVLLRSRILAHVRPQGWLPWNASNFGLRTSYFAEYQSSGPGALPSSRVAWSKQIKTVTDANKYQASVFIQG | |||
| Full Sequence |
|---|
| Protein Sequence Length: 602 Download |
| MVWNSREDEI ALYGSGSLES SKTSTRRKCT VTFVVLLLAG ILLAIIVPVT LHARNSSDDS 60 VPAQQAGPPA GVTTACSATQ YPDTCSNTLQ NSTHSDARIF TSTTVAAAST GVDETRLSIK 120 NSQTPENAPA VEVCLDTLTS ASAELEVVLK DLNTTDKAVL NATFDDIKTR LTAAMEFHTT 180 CLDALEEVGG PISPSVQAVS KKTNELFSVA LTFVNAFSAF GDDFLAWAKS FGMDLTQFNR 240 RGRRLLHDDL YSTGHEQDTV ETFPLWVGME QRRHLLQTVP ANGTYNVTVA WNGSGKYRKI 300 MDAVKNAPIK SSSPYVIYIK SGIYKEQVKI NSSLTNIMLL GDGPAYTIIT GSLSVALTKS 360 MTTFLSPTLI VEGQGFKAKG IQVRNTAGPA GHQAVALRVS ADKSSFYQCT FDSFQDTLYV 420 HNYRQFYRDC TIKGTIDYIF GNAWAVFQNC RLTAKKSTIV GQVNVYTAQG KTDRGQTTGI 480 SFQSCTFDAT TDLARNSKAF PTYLGRPWKA YATTVLLRSR ILAHVRPQGW LPWNASNFGL 540 RTSYFAEYQS SGPGALPSSR VAWSKQIKTV TDANKYQASV FIQGNSWVKA TNFPYTGALF 600 T* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02468 | PLN02468 | 4.0e-118 | 56 | 599 | 557 | + putative pectinesterase/pectinesterase inhibitor | ||
| PLN02301 | PLN02301 | 3.0e-119 | 67 | 599 | 550 | + pectinesterase/pectinesterase inhibitor | ||
| PLN02745 | PLN02745 | 3.0e-121 | 55 | 600 | 559 | + Putative pectinesterase/pectinesterase inhibitor | ||
| PLN02314 | PLN02314 | 3.0e-133 | 49 | 599 | 563 | + pectinesterase | ||
| pfam01095 | Pectinesterase | 6.0e-140 | 286 | 585 | 302 | + Pectinesterase. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004857 | enzyme inhibitor activity |
| GO:0005618 | cell wall |
| GO:0030599 | pectinesterase activity |
| GO:0042545 | cell wall modification |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| RefSeq | XP_001758305.1 | 0 | 76 | 594 | 2 | 493 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001759030.1 | 0 | 287 | 598 | 1 | 318 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001767596.1 | 0 | 69 | 599 | 13 | 532 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001767671.1 | 0 | 269 | 601 | 1 | 333 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001783825.1 | 0 | 132 | 596 | 23 | 461 | predicted protein [Physcomitrella patens subsp. patens] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1gq8_A | 0 | 286 | 599 | 8 | 319 | A Chain A, Pectin Methylesterase From Carrot |
| PDB | 1xg2_A | 0 | 286 | 600 | 4 | 316 | B Chain B, Crystal Structure Of The Complex Between Pectin Methylesterase And Its Inhibitor Protein |
| PDB | 1qjv_B | 2e-25 | 284 | 583 | 3 | 330 | A Chain A, Pectin Methylesterase Pema From Erwinia Chrysanthemi |
| PDB | 1qjv_A | 2e-25 | 284 | 583 | 3 | 330 | A Chain A, Pectin Methylesterase Pema From Erwinia Chrysanthemi |
| PDB | 2ntq_B | 8e-25 | 284 | 583 | 3 | 330 | A Chain A, Pectin Methylesterase Pema From Erwinia Chrysanthemi |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| homogalacturonan degradation | RXN-2102 | EC-3.1.1.11 | pectinesterase |
| Transmembrane Domains | ||||
|---|---|---|---|---|
 | ||||
| Start | End | |||
| 31 | 53 | |||
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
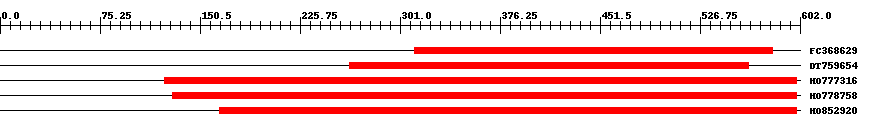 | ||||
| Hit | Length | Start | End | EValue |
| FC368629 | 270 | 312 | 581 | 0 |
| DT759654 | 301 | 263 | 563 | 0 |
| HO777316 | 480 | 124 | 599 | 0 |
| HO778758 | 472 | 130 | 599 | 0 |
| HO852920 | 437 | 165 | 599 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |