

| Basic Information | |
|---|---|
| Species | Physcomitrella patens |
| Cazyme ID | Pp1s378_15V6.1 |
| Family | CE8 |
| Protein Properties | Length: 577 Molecular Weight: 63561.2 Isoelectric Point: 8.6176 |
| Chromosome | Chromosome/Scaffold: 378 Start: 84023 End: 86564 |
| Description | Plant invertase/pectin methylesterase inhibitor superfamily |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CE8 | 259 | 560 | 0 |
| NVIVAKDGSGKYRTVGEAIMKAPKTGDKYAKRYVIYVKAGVYDEQIIIPKKLTNLMIIGDGIDKTIFTGSRNVALMKGMTTYLSGTMIVQGDGFVGRLFT CRNTAGASGHQAVATRVTADKVAFHRVKFDGFQDTLYCHALRQFYRDCVVIGTVDFIFGNANAVFQNCQIIARKTTLRGQQNTYTAQGRVNKYQNTGLAF QDCNFDGTADLKRNVQYYPTYLGRPWKAYSVCVLLRPTIQAHLDPAGWLPWNTTNFGLYTSFFAEYKGKGPGSNRRSRVKWSHEIKNPRVANRYQAASFI NG | |||
| Full Sequence |
|---|
| Protein Sequence Length: 577 Download |
| MSNDDEKPMV GSYRPKSRRC CVVSVLSVSL VLILVLCIAI PLGVKNSKND DGDNGVNVAC 60 QATEYPEACR ESVAGSDGTP VGMSRSSVKS STDELSYLQA NTTDEQCKEI LADGVAAFQR 120 VLVALETTNH TLVEETCADV QTDLSAVLTY VDTCKEMMQE SGSAEFHSFV QRALKSEQFT 180 GNSLALINGI CLYGGNLKNW AGKNFGVSMP DLDQFYGGGP SRRLMNADPY NDEGEEIQLP 240 SWMDSATSRH LLTRPASYNV IVAKDGSGKY RTVGEAIMKA PKTGDKYAKR YVIYVKAGVY 300 DEQIIIPKKL TNLMIIGDGI DKTIFTGSRN VALMKGMTTY LSGTMIVQGD GFVGRLFTCR 360 NTAGASGHQA VATRVTADKV AFHRVKFDGF QDTLYCHALR QFYRDCVVIG TVDFIFGNAN 420 AVFQNCQIIA RKTTLRGQQN TYTAQGRVNK YQNTGLAFQD CNFDGTADLK RNVQYYPTYL 480 GRPWKAYSVC VLLRPTIQAH LDPAGWLPWN TTNFGLYTSF FAEYKGKGPG SNRRSRVKWS 540 HEIKNPRVAN RYQAASFING KSWITNIGMP YTLQPV* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PLN02301 | PLN02301 | 2.0e-118 | 1 | 572 | 599 | + pectinesterase/pectinesterase inhibitor | ||
| PLN02314 | PLN02314 | 2.0e-121 | 12 | 571 | 597 | + pectinesterase | ||
| PLN02468 | PLN02468 | 6.0e-123 | 45 | 571 | 560 | + putative pectinesterase/pectinesterase inhibitor | ||
| PLN02745 | PLN02745 | 5.0e-123 | 52 | 573 | 556 | + Putative pectinesterase/pectinesterase inhibitor | ||
| pfam01095 | Pectinesterase | 7.0e-147 | 259 | 560 | 304 | + Pectinesterase. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004857 | enzyme inhibitor activity |
| GO:0005618 | cell wall |
| GO:0030599 | pectinesterase activity |
| GO:0042545 | cell wall modification |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| RefSeq | XP_001758305.1 | 0 | 60 | 570 | 2 | 493 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001767596.1 | 0 | 55 | 572 | 15 | 529 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001767671.1 | 0 | 249 | 572 | 5 | 328 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001772362.1 | 0 | 231 | 572 | 8 | 346 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_001783825.1 | 0 | 83 | 576 | 1 | 465 | predicted protein [Physcomitrella patens subsp. patens] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1xg2_A | 0 | 259 | 571 | 4 | 311 | B Chain B, Crystal Structure Of The Complex Between Pectin Methylesterase And Its Inhibitor Protein |
| PDB | 1gq8_A | 0 | 259 | 573 | 8 | 317 | A Chain A, Pectin Methylesterase From Carrot |
| PDB | 1qjv_B | 8e-21 | 257 | 531 | 3 | 311 | A Chain A, Pectin Methylesterase Pema From Erwinia Chrysanthemi |
| PDB | 1qjv_A | 8e-21 | 257 | 531 | 3 | 311 | A Chain A, Pectin Methylesterase Pema From Erwinia Chrysanthemi |
| PDB | 2ntq_B | 8e-21 | 257 | 531 | 3 | 311 | A Chain A, Pectin Methylesterase Pema From Erwinia Chrysanthemi |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| homogalacturonan degradation | RXN-2102 | EC-3.1.1.11 | pectinesterase |
| Transmembrane Domains | ||||
|---|---|---|---|---|
 | ||||
| Start | End | |||
| 21 | 43 | |||
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
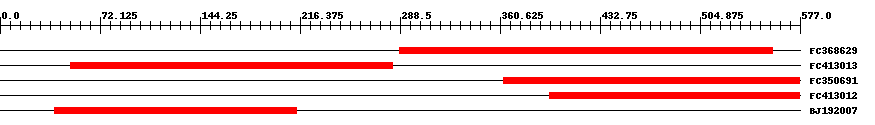 | ||||
| Hit | Length | Start | End | EValue |
| FC368629 | 270 | 288 | 557 | 0 |
| FC413013 | 233 | 51 | 283 | 0 |
| FC350691 | 215 | 363 | 577 | 0 |
| FC413012 | 182 | 396 | 577 | 0 |
| BJ192007 | 176 | 39 | 214 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |