

| Basic Information | |
|---|---|
| Species | Brassica rapa |
| Cazyme ID | Bra019249 |
| Family | GH37 |
| Protein Properties | Length: 556 Molecular Weight: 63177.8 Isoelectric Point: 5.8863 |
| Chromosome | Chromosome/Scaffold: 03 Start: 25445970 End: 25448554 |
| Description | trehalase 1 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH37 | 36 | 533 | 0 |
| PDPKFYIDLSLKLPHHLSTVEAAFNDLTSGSRDLPVPVKKLEKFVHEYFDDAKDLVPHEPEDFVTDPFEFLLNVENDQVREWAREVHSLWKTLCYRVSDS VRESPDRHTLLPLPEPVIIPGSRFKEVYYWDSYWVIKGLMTSKMFTTAKGIVTNLMSLVETYGYALNGARAYYTNRSQPPLLSSMVYEIYNVTKDEELVR KAIPVLLKEYEFWNSGKHKVVIRDASGNDHMLSRYYAMWNMPRPESCVFDQESASAFSSTLQKQRFLRDIATAAETGCDFSTRWMRDPPNFATMATTSVV PVDLNVFLLKMELDIAFMMGICGDKNGSERFVKASEARKKAFEAVFWNEKAGQWLDYWLSSDGDEPETWKAENQNTNVFASNFAPIWISSFNSDENLVKK VVKALKNSGLIAPAGILTSLKNSGQQWDYPNGWAPQQEIIVTGLARTGVKEAKEIAEEIARRWIRSSYSVYKTSGSIHEKLNVAEFGEYGGGGEYKPQ | |||
| Full Sequence |
|---|
| Protein Sequence Length: 556 Download |
| MAISDTGPLV QTTPLVTFLQ RVQLEAHRSY PNEKTPDPKF YIDLSLKLPH HLSTVEAAFN 60 DLTSGSRDLP VPVKKLEKFV HEYFDDAKDL VPHEPEDFVT DPFEFLLNVE NDQVREWARE 120 VHSLWKTLCY RVSDSVRESP DRHTLLPLPE PVIIPGSRFK EVYYWDSYWV IKGLMTSKMF 180 TTAKGIVTNL MSLVETYGYA LNGARAYYTN RSQPPLLSSM VYEIYNVTKD EELVRKAIPV 240 LLKEYEFWNS GKHKVVIRDA SGNDHMLSRY YAMWNMPRPE SCVFDQESAS AFSSTLQKQR 300 FLRDIATAAE TGCDFSTRWM RDPPNFATMA TTSVVPVDLN VFLLKMELDI AFMMGICGDK 360 NGSERFVKAS EARKKAFEAV FWNEKAGQWL DYWLSSDGDE PETWKAENQN TNVFASNFAP 420 IWISSFNSDE NLVKKVVKAL KNSGLIAPAG ILTSLKNSGQ QWDYPNGWAP QQEIIVTGLA 480 RTGVKEAKEI AEEIARRWIR SSYSVYKTSG SIHEKLNVAE FGEYGGGGEY KPQVDGLWVV 540 KRSYISILGG VWMAL* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| PRK13272 | treA | 4.0e-65 | 98 | 483 | 401 | + trehalase; Provisional | ||
| PRK13271 | treA | 6.0e-69 | 124 | 483 | 368 | + trehalase; Provisional | ||
| COG1626 | TreA | 7.0e-72 | 135 | 483 | 360 | + Neutral trehalase [Carbohydrate transport and metabolism] | ||
| pfam01204 | Trehalase | 2.0e-159 | 36 | 520 | 492 | + Trehalase. Trehalase (EC:3.2.1.28) is known to recycle trehalose to glucose. Trehalose is a physiological hallmark of heat-shock response in yeast and protects of proteins and membranes against a variety of stresses. This family is found in conjunction with pfam07492 in fungi. | ||
| PLN02567 | PLN02567 | 0 | 9 | 534 | 530 | + alpha,alpha-trehalase | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0004555 | alpha,alpha-trehalase activity |
| GO:0005991 | trehalose metabolic process |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAB63620.1 | 0 | 4 | 538 | 8 | 535 | trehalase precusor isolog [Arabidopsis thaliana] |
| GenBank | AAF22127.1 | 0 | 4 | 538 | 8 | 544 | AF126425_1 trehalase [Arabidopsis thaliana] |
| RefSeq | NP_194135.1 | 0 | 4 | 538 | 69 | 604 | TRE1 (TREHALASE 1); alpha,alpha-trehalase/ trehalase [Arabidopsis thaliana] |
| RefSeq | XP_002303675.1 | 0 | 7 | 538 | 15 | 546 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002518875.1 | 0 | 5 | 538 | 12 | 545 | alpha,alpha-trehalase, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2wyn_D | 0 | 79 | 537 | 58 | 488 | A Chain A, Characterization And Engineering Of The Bifunctional N- And O-glucosyltransferase Involved In Xenobiotic Metabolism In Plants |
| PDB | 2wyn_C | 0 | 79 | 537 | 58 | 488 | A Chain A, Characterization And Engineering Of The Bifunctional N- And O-glucosyltransferase Involved In Xenobiotic Metabolism In Plants |
| PDB | 2wyn_B | 0 | 79 | 537 | 58 | 488 | A Chain A, Characterization And Engineering Of The Bifunctional N- And O-glucosyltransferase Involved In Xenobiotic Metabolism In Plants |
| PDB | 2wyn_A | 0 | 79 | 537 | 58 | 488 | A Chain A, Characterization And Engineering Of The Bifunctional N- And O-glucosyltransferase Involved In Xenobiotic Metabolism In Plants |
| PDB | 2jjb_D | 0 | 79 | 537 | 58 | 488 | A Chain A, Characterization And Engineering Of The Bifunctional N- And O-glucosyltransferase Involved In Xenobiotic Metabolism In Plants |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| trehalose degradation II (trehalase) | TREHALA-RXN | EC-3.2.1.28 | α,α-trehalase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
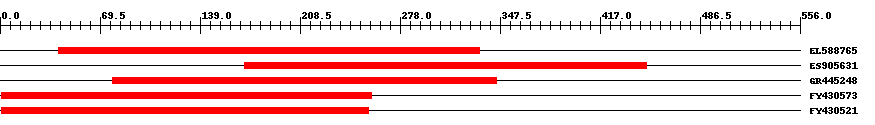 | ||||
| Hit | Length | Start | End | EValue |
| EL588765 | 293 | 41 | 333 | 0 |
| ES905631 | 280 | 170 | 449 | 0 |
| GR445248 | 269 | 78 | 345 | 0 |
| FY430573 | 258 | 1 | 258 | 0 |
| FY430521 | 256 | 1 | 256 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |