

| Basic Information | |
|---|---|
| Species | Chlamydomonas reinhardtii |
| Cazyme ID | g11709.t1 |
| Family | GH20 |
| Protein Properties | Length: 1248 Molecular Weight: 126231 Isoelectric Point: 5.4154 |
| Chromosome | Chromosome/Scaffold: 11 Start: 1680981 End: 1692262 |
| Description | beta-hexosaminidase 2 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH20 | 231 | 443 | 0 |
| SDWPRFPHRGLLLDTARHFLPLPALRAAVVALAAVKMNTLHWHAVDDQSFPLAAVAEAAGLPDLAARGAFGPRMSYSRQDVESLVVYAAARGVRVLLELD TPGHARSWGAGLPGLLSDCGGAGGGGSAGSGGGTTGTIDPTKDSSYAAVGSVLAAAAALLPERLLHLGGDEVDVGCWAADPAVRDWMARHGLSGDDEGAY LALQAHYMGRVLR | |||
| Full Sequence |
|---|
| Protein Sequence Length: 1248 Download |
| MRRTPILALL ALQAALASAV PAAELGLWPK PQRLVVTDET LALDPLGLPV SCKPAPCGQV 60 VLMAVARFRK NARLSHYRPA TWAAPAGATA GATATAGAPA ATAATAAAAV TELVLHVKDQ 120 SAPLQLGVDE AYDIQVPGGG GGSVHIRAAT QWGALHALET LSQLVVEVEG PAVAAGAGAA 180 VAAVVAVAEG AAAAAAGGRK AALQPKGNKG ELLQPHEMLQ PQLALVVANV SDWPRFPHRG 240 LLLDTARHFL PLPALRAAVV ALAAVKMNTL HWHAVDDQSF PLAAVAEAAG LPDLAARGAF 300 GPRMSYSRQD VESLVVYAAA RGVRVLLELD TPGHARSWGA GLPGLLSDCG GAGGGGSAGS 360 GGGTTGTIDP TKDSSYAAVG SVLAAAAALL PERLLHLGGD EVDVGCWAAD PAVRDWMARH 420 GLSGDDEGAY LALQAHYMGR VLRDVVSGRG SGAAGGAAAA AATAAATASA TASDPASGSG 480 SGVGGGGGGG GGAPPRTPVV WQEAFDVAGG AQLPRETIVQ VWKSDRGPTS RRSPGADAAA 540 TTAAAATAAT TAAAATAATT AAAATAITLA AAATAASTAA AATADAQQAR LREAAAAAAA 600 AAAVAAAGGW TSSSSSASLT DPQADVAGWA LEAEVEMEAA AAAGVEAALA AAVAADPGLD 660 EDEREELAEM LEAEAFAQEW VQQRRLLFDW SNLWGGGGGG GGGNNNAAER TGGHGGGSGG 720 AGGGGGMMRG GGLRAGGGGG GGAAGGGELG EGGMPVRRKK KMKKKKKMKK RKIVPAGGGG 780 GGDGYGGGGA DDDDDTGAWR RRTLLGQRLR FAHPEDANEA ERPSLSLARP YLHPGPHRAI 840 TVPRHRQEQE EQEGKKQQQQ QVEVEVEVAA VMPVEQQQME VEKEQQQQQQ QQKAAAVLRH 900 RGIGSGSGSS SSAASSSSSD TASRFYTGAT TRQHHAQLRT QQTHTQTDPQ QQQQVQQQGQ 960 QQQQQGEKQQ EGDQQEADAA AHAAAGAAAD LQAATTGPAT TAAAGDTAAV AAAAAAAQDT 1020 HAAAGTADHD KAHGDGGGNS NTGYDSGGDV DTRRALAALR GAVTAARAAR LGERGQGEEL 1080 AAVTAAGFRA IVSSGWYLDW VSWGQDWRRY WAQEPLAGLG SAAQRALVIG GEACMWGEYV 1140 DATNLMSRTW PRASAVAERL WSAPLEPDNN DNNDDNDNNT QRQQQQRQQQ QQQQQEAEED 1200 AAARIAVHRC RLLARGIPAQ PLAPGACPND GDFDDVSSGS REEEQAE* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
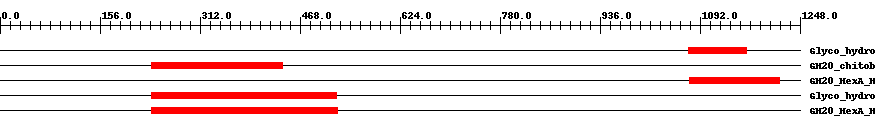 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam00728 | Glyco_hydro_20 | 3.0e-16 | 1074 | 1164 | 107 | + Glycosyl hydrolase family 20, catalytic domain. This domain has a TIM barrel fold. | ||
| cd06570 | GH20_chitobiase-like_1 | 3.0e-28 | 236 | 441 | 206 | + A functionally uncharacterized subgroup of the Glycosyl hydrolase family 20 (GH20) catalytic domain found in proteins similar to the chitobiase of Serratia marcescens, a beta-N-1,4-acetylhexosaminidase that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. This subgroup lacks the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. | ||
| cd06562 | GH20_HexA_HexB-like | 3.0e-39 | 1075 | 1216 | 149 | + Beta-N-acetylhexosaminidases catalyze the removal of beta-1,4-linked N-acetyl-D-hexosamine residues from the non-reducing ends of N-acetyl-beta-D-hexosaminides including N-acetylglucosides and N-acetylgalactosides. The hexA and hexB genes encode the alpha- and beta-subunits of the two major beta-N-acetylhexosaminidase isoenzymes, N-acetyl-beta-D-hexosaminidase A (HexA) and beta-N-acetylhexosaminidase B (HexB). Both the alpha and the beta catalytic subunits have a TIM-barrel fold and belong to the glycosyl hydrolase family 20 (GH20). The HexA enzyme is a heterodimer containing one alpha and one beta subunit while the HexB enzyme is a homodimer containing two beta-subunits. Hexosaminidase mutations cause an inability to properly hydrolyze certain sphingolipids which accumulate in lysosomes within the brain, resulting in the lipid storage disorders Tay-Sachs and Sandhoff. Mutations in the alpha subunit cause in a deficiency in the HexA enzyme and result in Tay-Sachs, mutations in the beta-subunit cause in a deficiency in both HexA and HexB enzymes and result in Sandhoff disease. In both disorders GM(2) gangliosides accumulate in lysosomes. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. | ||
| pfam00728 | Glyco_hydro_20 | 8.0e-42 | 236 | 525 | 290 | + Glycosyl hydrolase family 20, catalytic domain. This domain has a TIM barrel fold. | ||
| cd06562 | GH20_HexA_HexB-like | 3.0e-60 | 236 | 526 | 291 | + Beta-N-acetylhexosaminidases catalyze the removal of beta-1,4-linked N-acetyl-D-hexosamine residues from the non-reducing ends of N-acetyl-beta-D-hexosaminides including N-acetylglucosides and N-acetylgalactosides. The hexA and hexB genes encode the alpha- and beta-subunits of the two major beta-N-acetylhexosaminidase isoenzymes, N-acetyl-beta-D-hexosaminidase A (HexA) and beta-N-acetylhexosaminidase B (HexB). Both the alpha and the beta catalytic subunits have a TIM-barrel fold and belong to the glycosyl hydrolase family 20 (GH20). The HexA enzyme is a heterodimer containing one alpha and one beta subunit while the HexB enzyme is a homodimer containing two beta-subunits. Hexosaminidase mutations cause an inability to properly hydrolyze certain sphingolipids which accumulate in lysosomes within the brain, resulting in the lipid storage disorders Tay-Sachs and Sandhoff. Mutations in the alpha subunit cause in a deficiency in the HexA enzyme and result in Tay-Sachs, mutations in the beta-subunit cause in a deficiency in both HexA and HexB enzymes and result in Sandhoff disease. In both disorders GM(2) gangliosides accumulate in lysosomes. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. | ||
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2GK1 | 0 | 27 | 525 | 1 | 373 | B Chain B, X-Ray Crystal Structure Of Ngt-Bound Hexa |
| PDB | 2GK1 | 7e-31 | 1078 | 1227 | 380 | 500 | B Chain B, X-Ray Crystal Structure Of Ngt-Bound Hexa |
| GenBank | AAA51827.1 | 0 | 146 | 525 | 88 | 356 | N-acetyl-alpha-glucosaminidase prepro-polypeptide [Homo sapiens] |
| GenBank | AAA51827.1 | 3e-31 | 1078 | 1227 | 363 | 483 | N-acetyl-alpha-glucosaminidase prepro-polypeptide [Homo sapiens] |
| RefSeq | NP_000511.2 | 0 | 27 | 525 | 23 | 395 | beta-hexosaminidase subunit alpha preproprotein [Homo sapiens] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2gk1_G | 0 | 27 | 525 | 1 | 373 | B Chain B, X-Ray Crystal Structure Of Ngt-Bound Hexa |
| PDB | 2gk1_G | 1e-32 | 1078 | 1227 | 380 | 500 | B Chain B, X-Ray Crystal Structure Of Ngt-Bound Hexa |
| PDB | 2gk1_E | 0 | 27 | 525 | 1 | 373 | B Chain B, X-Ray Crystal Structure Of Ngt-Bound Hexa |
| PDB | 2gk1_E | 1e-32 | 1078 | 1227 | 380 | 500 | B Chain B, X-Ray Crystal Structure Of Ngt-Bound Hexa |
| PDB | 2gk1_C | 0 | 27 | 525 | 1 | 373 | B Chain B, X-Ray Crystal Structure Of Ngt-Bound Hexa |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| chitin degradation II | RXN-12625 | EC-3.2.1.52 | β-L-N-acetylhexosaminidase |
| chitin degradation II | RXN-12626 | EC-3.2.1.52 | β-L-N-acetylhexosaminidase |