

| Basic Information | |
|---|---|
| Species | Manihot esculenta |
| Cazyme ID | cassava4.1_029798m |
| Family | CBM20 |
| Protein Properties | Length: 376 Molecular Weight: 41756.3 Isoelectric Point: 4.2242 |
| Chromosome | Chromosome/Scaffold: 09086 Start: 8682 End: 10413 |
| Description | Carbohydrate-binding-like fold |
| View CDS | |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM20 | 89 | 175 | 1.4e-23 |
| VHVKFHLNKECSFGEQFALVGDDQMFGMWDPENAIPLNWSEGHVWTLELDIPIGKFVHFKFILREITGEILWQPGPDRILKTWETKN | |||
| Full Sequence |
|---|
| Protein Sequence Length: 376 Download |
| MKALPISSSW SKISFQRNAD CPAFNPTGHQ VRCFQSKNFL NVGFLRCVKQ KPLLSVSAST 60 SPLSPDSETA TDLEEAQSKT QESYQPNTVH VKFHLNKECS FGEQFALVGD DQMFGMWDPE 120 NAIPLNWSEG HVWTLELDIP IGKFVHFKFI LREITGEILW QPGPDRILKT WETKNTIVVS 180 EDWEDAAFQQ VIEEGPIAGE PTGNSEMLFV ADKLTTNSEM LVFAENLTHP KGDLVSDGND 240 YPANEQQPSN HEKPITVDNI PQPEETDTKE DPVEGINHEK DENKANLNKE AMTAGMNLAS 300 MNVEGNLVDA HEGDPFLVPG LPPSSAFPND PVTHDEGETS LAFGASVVND EVKNHNMPEL 360 VEKHEVVGDP HQEEET |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
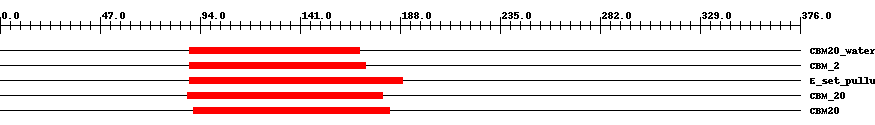 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| cd05818 | CBM20_water_dikinase | 6.0e-11 | 89 | 169 | 81 | + Phosphoglucan water dikinase (also known as alpha-glucan water dikinase), N-terminal CBM20 (carbohydrate-binding module, family 20) domain. This domain is found in the chloroplast-encoded phosphoglucan water dikinase, one of two enzymes involved in the phosphorylation of plant starches. In addition to the CBM20 domain, phosphoglucan water dikinase contains a C-terminal pyruvate binding domain. The CBM20 domain is found in a large number of starch degrading enzymes including alpha-amylase, beta-amylase, glucoamylase, and CGTase (cyclodextrin glucanotransferase). CBM20 is also present in proteins that have a regulatory role in starch metabolism in plants (e.g. alpha-amylase) or glycogen metabolism in mammals (e.g. laforin). CBM20 folds as an antiparallel beta-barrel structure with two starch binding sites. These two sites are thought to differ functionally with site 1 acting as the initial starch recognition site and site 2 involved in the specific recognition of appropriate regions of starch. | ||
| smart01065 | CBM_2 | 2.0e-14 | 89 | 172 | 88 | + Starch binding domain. | ||
| cd02861 | E_set_pullulanase_like | 2.0e-15 | 89 | 189 | 103 | + Early set domain associated with the catalytic domain of pullulanase-like proteins. E or "early" set domains are associated with the catalytic domain of pullulanase at either the N-terminal or C-terminal end, and in a few instances at both ends. Pullulanase (also called dextrinase or alpha-dextrin endo-1,6-alpha glucosidase) is an enzyme with action similar to that of isoamylase; it cleaves 1,6-alpha-glucosidic linkages in pullulan, amylopectin, and glycogen, and in alpha-and beta-amylase limit-dextrins of amylopectin and glycogen. The E set domain of pullulanase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase. This domain is also a member of the CBM48 (Carbohydrate Binding Module 48) family whose members include maltooligosyl trehalose synthase, starch branching enzyme, glycogen branching enzyme, glycogen debranching enzyme, isoamylase, and the beta subunit of AMP-activated protein kinase. | ||
| pfam00686 | CBM_20 | 4.0e-16 | 88 | 180 | 96 | + Starch binding domain. | ||
| cd05467 | CBM20 | 5.0e-24 | 91 | 183 | 96 | + The family 20 carbohydrate-binding module (CBM20), also known as the starch-binding domain, is found in a large number of starch degrading enzymes including alpha-amylase, beta-amylase, glucoamylase, and CGTase (cyclodextrin glucanotransferase). CBM20 is also present in proteins that have a regulatory role in starch metabolism in plants (e.g. alpha-amylase) or glycogen metabolism in mammals (e.g. laforin). CBM20 folds as an antiparallel beta-barrel structure with two starch binding sites. These two sites are thought to differ functionally with site 1 acting as the initial starch recognition site and site 2 involved in the specific recognition of appropriate regions of starch. | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:2001070 | starch binding |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| EMBL | CAN60108.1 | 0 | 30 | 376 | 4 | 369 | hypothetical protein [Vitis vinifera] |
| EMBL | CBI30404.1 | 0 | 30 | 376 | 28 | 312 | unnamed protein product [Vitis vinifera] |
| RefSeq | XP_002308216.1 | 0 | 85 | 376 | 26 | 333 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002322974.1 | 0 | 1 | 260 | 1 | 268 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002528119.1 | 0 | 85 | 360 | 16 | 321 | catalytic, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1kum_A | 0.00003 | 86 | 166 | 4 | 88 | A Chain A, Crystal Structure Of Brassica Juncea Chitinase Catalytic Module (Bjchi3) |
| PDB | 1kul_A | 0.00003 | 86 | 166 | 4 | 88 | A Chain A, Crystal Structure Of Brassica Juncea Chitinase Catalytic Module (Bjchi3) |
| PDB | 1acz_A | 0.00003 | 86 | 166 | 4 | 88 | A Chain A, Glucoamylase, Granular Starch-Binding Domain Complex With Cyclodextrin, Nmr, 5 Structures |
| PDB | 1ac0_A | 0.00003 | 86 | 166 | 4 | 88 | A Chain A, Glucoamylase, Granular Starch-Binding Domain Complex With Cyclodextrin, Nmr, Minimized Average Structure |
| PDB | 1cyg_A | 0.00003 | 89 | 185 | 580 | 680 | A Chain A, Cyclodextrin Glucanotransferase (E.C.2.4.1.19) (Cgtase) |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
 | ||||
| Hit | Length | Start | End | EValue |
| DB927622 | 141 | 1 | 141 | 0 |
| DV449539 | 125 | 236 | 360 | 0 |
| GO924256 | 267 | 31 | 286 | 0 |
| FQ344867 | 232 | 1 | 230 | 0 |
| FC860701 | 203 | 33 | 230 | 0 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |