

| Basic Information | |
|---|---|
| Species | Glycine max |
| Cazyme ID | Glyma03g27600.1 |
| Family | CBM20 |
| Protein Properties | Length: 966 Molecular Weight: 111608 Isoelectric Point: 5.9847 |
| Chromosome | Chromosome/Scaffold: 03 Start: 35337374 End: 35349555 |
| Description | disproportionating enzyme 2 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM20 | 16 | 103 | 1.1e-22 |
| VKVSFRIPYFTQWGQTLLVCGSVPVLGSWNVKKGVLLRPIHQGAELIWGGSITVPKGFRCQYSYYVVDDNKNVLRWEMGKKHELVLPE | |||
| GH77 | 272 | 920 | 0 |
| IPMFSIRSESDLGVGEFLDLKLLVDWAVATGFHLVQLLPINDTSVHGMWWDSYPYSSLSVFALHPLYLRVQALSKNIPEAIKKEIEKAKQQLDGKDVDYE ATMATKLSIAKKVFAQEKDLILNSSSFKEFFSENEGWLKPYAAFCFLRDFFETSDRTQWGHFAHYSEDKLEKLVSKDSLHYEIICFHYYVQYHLHLQLSE AAEYARKKGVILKGDLPIGVDRNSVDTWVYPNLFRMNTSTGAPPDYFDKNGQNWGFPTYNWEEMSKDNYGWWRARLTQMAKYFTAYRIDHILGFFRIWEL PDHAATGLVGKFRPSIPLSLEELEREGIWDFNRLSRPYIKRELLQEKFGDAWTFVATTFLNEIDKNFYEFKEDCNTEKKIASKLKICAESSLLLESVDKL RHNLFDLSQNIVLIRDSEDPRKFYPRFNLEDTSSFQDLDDHSKNVLKRLYNDYYFCRQENLWRQNALKTLPVLLNSSDMLACGEDLGLIPSCVHPVMQEL GLVGLRIQRMPNEPDLEFGIPSKYSYMTVCAPSCHDCSTLRAWWEEDEERRLRFFKNVMESDELPPDQCVPEVVHFVLRQHFEAPSMWAIFPLQDLLALK EEYTTRPATEETINDPTNPKHYWRYRVHVTLESLIKDNDLQTAIKDLVR | |||
| Full Sequence |
|---|
| Protein Sequence Length: 966 Download |
| MVNPGLFSAN KSVNSVKVSF RIPYFTQWGQ TLLVCGSVPV LGSWNVKKGV LLRPIHQGAE 60 LIWGGSITVP KGFRCQYSYY VVDDNKNVLR WEMGKKHELV LPEGIRSGHE IEFRDLWQTG 120 SDALPFRSAF KDVIFRQCWD LSDTTVGVNH INIEPEGEAI LVQFKISCPN IEKDTSIYVI 180 GSNTKLGQWK VENGLKLSYF GESVWKSECV MQRSDFPIKY RYGKYDRCGN FSIESGPNRE 240 VSTNSSRSEA KYIFLSDGMM REIPWRGAGV AIPMFSIRSE SDLGVGEFLD LKLLVDWAVA 300 TGFHLVQLLP INDTSVHGMW WDSYPYSSLS VFALHPLYLR VQALSKNIPE AIKKEIEKAK 360 QQLDGKDVDY EATMATKLSI AKKVFAQEKD LILNSSSFKE FFSENEGWLK PYAAFCFLRD 420 FFETSDRTQW GHFAHYSEDK LEKLVSKDSL HYEIICFHYY VQYHLHLQLS EAAEYARKKG 480 VILKGDLPIG VDRNSVDTWV YPNLFRMNTS TGAPPDYFDK NGQNWGFPTY NWEEMSKDNY 540 GWWRARLTQM AKYFTAYRID HILGFFRIWE LPDHAATGLV GKFRPSIPLS LEELEREGIW 600 DFNRLSRPYI KRELLQEKFG DAWTFVATTF LNEIDKNFYE FKEDCNTEKK IASKLKICAE 660 SSLLLESVDK LRHNLFDLSQ NIVLIRDSED PRKFYPRFNL EDTSSFQDLD DHSKNVLKRL 720 YNDYYFCRQE NLWRQNALKT LPVLLNSSDM LACGEDLGLI PSCVHPVMQE LGLVGLRIQR 780 MPNEPDLEFG IPSKYSYMTV CAPSCHDCST LRAWWEEDEE RRLRFFKNVM ESDELPPDQC 840 VPEVVHFVLR QHFEAPSMWA IFPLQDLLAL KEEYTTRPAT EETINDPTNP KHYWRYRVHV 900 TLESLIKDND LQTAIKDLVR WSGRSLPKED DSEVEVSPVS ALSSAEALSE KQQFAGTMEK 960 PVLVK* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam02446 | Glyco_hydro_77 | 9.0e-47 | 743 | 920 | 179 | + 4-alpha-glucanotransferase. These enzymes EC:2.4.1.25 transfer a segment of a (1,4)-alpha-D-glucan to a new 4-position in an acceptor, which may be glucose or (1,4)-alpha-D-glucan. | ||
| COG1640 | MalQ | 1.0e-67 | 269 | 573 | 307 | + 4-alpha-glucanotransferase [Carbohydrate transport and metabolism] | ||
| pfam02446 | Glyco_hydro_77 | 2.0e-108 | 272 | 582 | 313 | + 4-alpha-glucanotransferase. These enzymes EC:2.4.1.25 transfer a segment of a (1,4)-alpha-D-glucan to a new 4-position in an acceptor, which may be glucose or (1,4)-alpha-D-glucan. | ||
| PLN02950 | PLN02950 | 0 | 20 | 915 | 898 | + 4-alpha-glucanotransferase | ||
| PLN03236 | PLN03236 | 0 | 237 | 925 | 716 | + 4-alpha-glucanotransferase; Provisional | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0003824 | catalytic activity |
| GO:0004134 | 4-alpha-glucanotransferase activity |
| GO:0005975 | carbohydrate metabolic process |
| GO:2001070 | starch binding |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | AAR99599.1 | 0 | 1 | 930 | 1 | 922 | 4-alpha-glucanotransferase [Solanum tuberosum] |
| EMBL | CBI32836.1 | 0 | 1 | 960 | 1 | 949 | unnamed protein product [Vitis vinifera] |
| RefSeq | XP_002278329.1 | 0 | 1 | 960 | 1 | 949 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002308854.1 | 0 | 1 | 933 | 1 | 948 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002323208.1 | 0 | 1 | 933 | 6 | 898 | predicted protein [Populus trichocarpa] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 1tz7_B | 2e-27 | 266 | 583 | 22 | 329 | A Chain A, Aquifex Aeolicus Amylomaltase |
| PDB | 1tz7_A | 2e-27 | 266 | 583 | 22 | 329 | A Chain A, Aquifex Aeolicus Amylomaltase |
| PDB | 2x1i_A | 4e-26 | 301 | 583 | 39 | 316 | A Chain A, Glycoside Hydrolase Family 77 4-Alpha-Glucanotransferase From Thermus Brockianus |
| PDB | 1esw_A | 2e-25 | 307 | 583 | 45 | 316 | A Chain A, Glycoside Hydrolase Family 77 4-Alpha-Glucanotransferase From Thermus Brockianus |
| Metabolic Pathways | |||
|---|---|---|---|
| Pathway Name | Reaction | EC | Protein Name |
| starch degradation I | RXN-1828 | EC-2.4.1.25 | 4-α-glucanotransferase |
| starch degradation II | RXN-12391 | EC-2.4.1.25 | 4-α-glucanotransferase |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
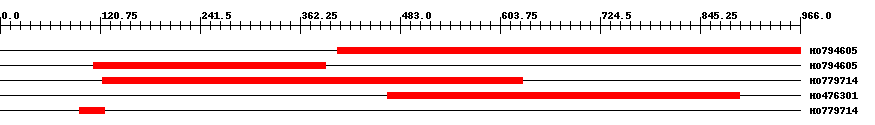 | ||||
| Hit | Length | Start | End | EValue |
| HO794605 | 568 | 407 | 966 | 0 |
| HO794605 | 281 | 113 | 393 | 0 |
| HO779714 | 508 | 124 | 631 | 0 |
| HO476301 | 431 | 468 | 893 | 0 |
| HO779714 | 31 | 96 | 126 | 1.8 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |