

| Basic Information | |
|---|---|
| Species | Phaseolus vulgaris |
| Cazyme ID | Phvul.006G183300.2 |
| Family | CBM57 |
| Protein Properties | Length: 636 Molecular Weight: 69647.3 Isoelectric Point: 9.2073 |
| Chromosome | Chromosome/Scaffold: 06 Start: 29236373 End: 29242359 |
| Description | receptor like protein 4 |
| View CDS | |
| External Links |
|---|
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM57 | 233 | 364 | 5.4e-24 |
| FGEDSDRPRSVETRIKQASHPPNFYPETLYRSALVSTSNQLDLTYTLDVDPNKNYSVWLHFAEIENSVTAAGQRVFDIMINGDVAFRNVDIVKLSGDRYT ALVLNKTVPVNGRTLTVTLSPIKGSFAIISAI | |||
| Full Sequence |
|---|
| Protein Sequence Length: 636 Download |
| MANIATFGFL LLWTLFLCFA FVSPAAQRGP YGMHISCGAR HDVQTKPTTT LWYKDFGYTG 60 GIPSNASTTS YIAPPLKTLR YFPLSEGLSN CYNINRVPKG RYSIRIFFGL VAQTRVTDEP 120 LFDISIQGTQ IYSLKSGWTS RDDQAFTEAQ VFLKDGSVSI CFHGTGHGDP AILSIEILQI 180 DNNAYYFGPD WSQGVILRTV KRLSCGFGQS KFDVDYGADP RGGDRYWQRI KTFGEDSDRP 240 RSVETRIKQA SHPPNFYPET LYRSALVSTS NQLDLTYTLD VDPNKNYSVW LHFAEIENSV 300 TAAGQRVFDI MINGDVAFRN VDIVKLSGDR YTALVLNKTV PVNGRTLTVT LSPIKGSFAI 360 ISAIEILEVI TAESKTLSDE VMALQTLKKA LGLPPRFGWN GDPCVPQQHP WTGADCRLDK 420 SSSKWVIDGL GLGNQGLKGF LPNDISRLHN LQILNLSGNG IRGAIPSPLG TITSLQVLDL 480 SYNFFNGSIP ESLGQLTSLQ RLNLNGNFLS GRVPATLGGR LLHGASFNFT DNAGLCGIPG 540 LPTCGPHLSA GAKVGIGLGA SFTFLLLIIG SVCWWKRRQN ILRVQQIAGR AAPYAKARTQ 600 FSRDIQMARH NNNTNNNNYG NAHTAVETGP ILLSR* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
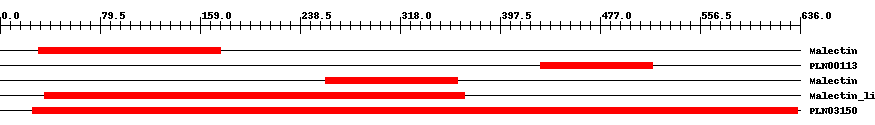 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam11721 | Malectin | 0.003 | 31 | 175 | 164 | + Di-glucose binding within endoplasmic reticulum. Malectin is a membrane-anchored protein of the endoplasmic reticulum that recognises and binds Glc2-N-glycan. It carries a signal peptide from residues 1-26, a C-terminal transmembrane helix from residues 255-274, and a highly conserved central part of approximately 190 residues followed by an acidic, glutamate-rich region. Carbohydrate-binding is mediated by the four aromatic residues, Y67, Y89, Y116, and F117 and the aspartate at D186. NMR-based ligand-screening studies has shown binding of the protein to maltose and related oligosaccharides, on the basis of which the protein has been designated "malectin", and its endogenous ligand is found to be Glc2-high-mannose N-glycan. | ||
| PLN00113 | PLN00113 | 6.0e-15 | 430 | 519 | 90 | + leucine-rich repeat receptor-like protein kinase; Provisional | ||
| pfam11721 | Malectin | 1.0e-17 | 259 | 364 | 118 | + Di-glucose binding within endoplasmic reticulum. Malectin is a membrane-anchored protein of the endoplasmic reticulum that recognises and binds Glc2-N-glycan. It carries a signal peptide from residues 1-26, a C-terminal transmembrane helix from residues 255-274, and a highly conserved central part of approximately 190 residues followed by an acidic, glutamate-rich region. Carbohydrate-binding is mediated by the four aromatic residues, Y67, Y89, Y116, and F117 and the aspartate at D186. NMR-based ligand-screening studies has shown binding of the protein to maltose and related oligosaccharides, on the basis of which the protein has been designated "malectin", and its endogenous ligand is found to be Glc2-high-mannose N-glycan. | ||
| pfam12819 | Malectin_like | 6.0e-67 | 35 | 369 | 354 | + Carbohydrate-binding protein of the ER. Malectin is a membrane-anchored protein of the endoplasmic reticulum that recognises and binds Glc2-N-glycan. The domain is found on a number of plant receptor kinases. | ||
| PLN03150 | PLN03150 | 0 | 26 | 634 | 609 | + hypothetical protein; Provisional | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005515 | protein binding |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| EMBL | CBI30598.1 | 0 | 24 | 634 | 18 | 624 | unnamed protein product [Vitis vinifera] |
| RefSeq | NP_174156.1 | 0 | 26 | 634 | 22 | 626 | AtRLP4 (Receptor Like Protein 4); protein binding [Arabidopsis thaliana] |
| RefSeq | XP_002279791.1 | 0 | 33 | 634 | 1 | 600 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002305781.1 | 0 | 33 | 634 | 1 | 596 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002509445.1 | 0 | 14 | 634 | 11 | 629 | serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3rgz_A | 0.00000000000006 | 437 | 551 | 644 | 755 | A Chain A, Structure Of The Starch Branching Enzyme I (bei) Complexed With Maltopentaose From Oryza Sativa L |
| PDB | 3rgz_A | 0.00000004 | 437 | 517 | 478 | 558 | A Chain A, Structure Of The Starch Branching Enzyme I (bei) Complexed With Maltopentaose From Oryza Sativa L |
| PDB | 3rgz_A | 0.0000004 | 424 | 517 | 393 | 486 | A Chain A, Structure Of The Starch Branching Enzyme I (bei) Complexed With Maltopentaose From Oryza Sativa L |
| PDB | 3rgz_A | 0.0000008 | 437 | 518 | 454 | 535 | A Chain A, Structure Of The Starch Branching Enzyme I (bei) Complexed With Maltopentaose From Oryza Sativa L |
| PDB | 3rgz_A | 0.00001 | 437 | 540 | 430 | 532 | A Chain A, Structure Of The Starch Branching Enzyme I (bei) Complexed With Maltopentaose From Oryza Sativa L |