

| Basic Information | |
|---|---|
| Species | Physcomitrella patens |
| Cazyme ID | Pp1s2_327V6.1 |
| Family | CBM57 |
| Protein Properties | Length: 643 Molecular Weight: 69906.1 Isoelectric Point: 6.1403 |
| Chromosome | Chromosome/Scaffold: 2 Start: 1654777 End: 1659509 |
| Description | receptor like protein 4 |
| View CDS | |
| External Links |
|---|
| NCBI Taxonomy |
| Plaza |
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| CBM57 | 238 | 376 | 3.5e-22 |
| RYWATDQTLFTARSDISLLNTSNNITKYSDPPNTYPQAIYQSATTTGPSNKLSYTIPVVPNQNYSVWLHFAEIDPSITGPNMRVFNVLANGQPLFEVVDI VKIAGAPFTALVLNKTVFVERKSFELTFEPLTGEIAVNA | |||
| Full Sequence |
|---|
| Protein Sequence Length: 643 Download |
| MLSVPSCRKL SLGSIMDRVH LTLFVVALSI FFRATSTAGQ SGAPIAVRIA CGSETESHAP 60 ESGYLWLRDM NYTGGSSGNL TTVSRIASQL DTVRYFELSD GNENCYNVSV PNGQYLIRFY 120 FSYGVKDNSG REPEFDVSLE GTLVYSLTRG WSTAVDYEYA DSTVRILDGA ATICFHSSGH 180 GNPVVASIEI LQIHDSAYNL GASSPSNVIL KTVKRVSAGA EKSGYGVSLH ADPWSGDRYW 240 ATDQTLFTAR SDISLLNTSN NITKYSDPPN TYPQAIYQSA TTTGPSNKLS YTIPVVPNQN 300 YSVWLHFAEI DPSITGPNMR VFNVLANGQP LFEVVDIVKI AGAPFTALVL NKTVFVERKS 360 FELTFEPLTG EIAVNAFEVF QLITREQPTA TDNVWALQSM KHSLSLPARL GWNGDPCVPQ 420 VHAWDGVNCE FDGAFGTWFI SGVDLNVQGL GGQLGDDLLG LQRLQTLNFS KNYLTGPIPE 480 SLGKISSLTT LDLSYNQLNG SIPPSLAQLS KLQELLLNDN QLSGEVPPAL GALPIRGATL 540 NLTNNDGLCG VGLQSCESVN GSRWTTFFLL VLGFLILVLG GYLCYKRRMN MLRGAQRLPR 600 DAPYAKARTT FVRDVQMARS VLANHFSRPA PSCTETAPLN PH* |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
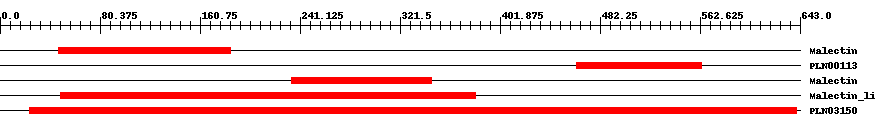 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam11721 | Malectin | 0.0007 | 47 | 185 | 157 | + Di-glucose binding within endoplasmic reticulum. Malectin is a membrane-anchored protein of the endoplasmic reticulum that recognises and binds Glc2-N-glycan. It carries a signal peptide from residues 1-26, a C-terminal transmembrane helix from residues 255-274, and a highly conserved central part of approximately 190 residues followed by an acidic, glutamate-rich region. Carbohydrate-binding is mediated by the four aromatic residues, Y67, Y89, Y116, and F117 and the aspartate at D186. NMR-based ligand-screening studies has shown binding of the protein to maltose and related oligosaccharides, on the basis of which the protein has been designated "malectin", and its endogenous ligand is found to be Glc2-high-mannose N-glycan. | ||
| PLN00113 | PLN00113 | 2.0e-9 | 463 | 564 | 108 | + leucine-rich repeat receptor-like protein kinase; Provisional | ||
| pfam11721 | Malectin | 6.0e-17 | 234 | 347 | 114 | + Di-glucose binding within endoplasmic reticulum. Malectin is a membrane-anchored protein of the endoplasmic reticulum that recognises and binds Glc2-N-glycan. It carries a signal peptide from residues 1-26, a C-terminal transmembrane helix from residues 255-274, and a highly conserved central part of approximately 190 residues followed by an acidic, glutamate-rich region. Carbohydrate-binding is mediated by the four aromatic residues, Y67, Y89, Y116, and F117 and the aspartate at D186. NMR-based ligand-screening studies has shown binding of the protein to maltose and related oligosaccharides, on the basis of which the protein has been designated "malectin", and its endogenous ligand is found to be Glc2-high-mannose N-glycan. | ||
| pfam12819 | Malectin_like | 6.0e-53 | 49 | 382 | 353 | + Carbohydrate-binding protein of the ER. Malectin is a membrane-anchored protein of the endoplasmic reticulum that recognises and binds Glc2-N-glycan. The domain is found on a number of plant receptor kinases. | ||
| PLN03150 | PLN03150 | 0 | 24 | 640 | 623 | + hypothetical protein; Provisional | ||
| Gene Ontology | |
|---|---|
| GO Term | Description |
| GO:0005515 | protein binding |
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| GenBank | ABH07898.1 | 0 | 47 | 625 | 1 | 586 | leucine-rich repeat family protein [Solanum lycopersicum] |
| EMBL | CBI30598.1 | 0 | 43 | 619 | 23 | 605 | unnamed protein product [Vitis vinifera] |
| RefSeq | XP_001752825.1 | 0 | 44 | 642 | 1 | 597 | predicted protein [Physcomitrella patens subsp. patens] |
| RefSeq | XP_002305781.1 | 0 | 47 | 638 | 1 | 592 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002509445.1 | 0 | 24 | 638 | 4 | 625 | serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 3rgz_A | 0.0000000006 | 443 | 564 | 637 | 756 | 1 Chain 1, Structure Of Bacillus Subtilis Pdaa In Complex With Nag, A Family 4 Carbohydrate Esterase. |
| PDB | 3rgz_A | 0.00000003 | 445 | 530 | 401 | 486 | 1 Chain 1, Structure Of Bacillus Subtilis Pdaa In Complex With Nag, A Family 4 Carbohydrate Esterase. |
| PDB | 3rgz_A | 0.00000004 | 464 | 560 | 396 | 493 | 1 Chain 1, Structure Of Bacillus Subtilis Pdaa In Complex With Nag, A Family 4 Carbohydrate Esterase. |
| PDB | 3rgz_A | 0.0000003 | 450 | 545 | 430 | 524 | 1 Chain 1, Structure Of Bacillus Subtilis Pdaa In Complex With Nag, A Family 4 Carbohydrate Esterase. |
| PDB | 3rgz_A | 0.00002 | 456 | 533 | 337 | 441 | 1 Chain 1, Structure Of Bacillus Subtilis Pdaa In Complex With Nag, A Family 4 Carbohydrate Esterase. |