

| Basic Information | |
|---|---|
| Species | Aquilegia coerulea |
| Cazyme ID | Aquca_002_00330.1 |
| Family | GH89 |
| Protein Properties | Length: 898 Molecular Weight: 102105 Isoelectric Point: 5.7745 |
| Chromosome | Chromosome/Scaffold: 2 Start: 2960255 End: 2972403 |
| Description | alpha-N-acetylglucosaminidase family / NAGLU family |
| View CDS | |
| External Links |
|---|
| CAZyDB |
| Signature Domain Download full data set without filtering | |||
|---|---|---|---|
 | |||
| Family | Start | End | Evalue |
| GH89 | 155 | 497 | 0 |
| PEIRIGGSSGVELSAGLHWYLKNWCGAHISWDKTGGAQLFSVPKPGFLPLVQASGVLIQRPVPWNYYQNAVASSYTFAWWDWERWEKEIDWMAVQGINLP LAFTGQEAIWQKVFQIFNISSSELNDFFGGPAFLAWSRMGNLHGWGGPLPQSWFDQQLSLQKKILGRMYELGMTPVLPAFSGNVPAALKSIYPSAKIARL GNWFTVRGDTRWCCTYLLDAMDPLFVEIGKAFVEMEITEYGRSSHIYNCDTFDENTPPVDDPEYISSLGAAIYRAMQSGDVDAVWLMQGWLFSYDPFWRP EQMKALLHSVPVGKLVVLDLFAEVKPLWITSEQFYGVPYIWKV | |||
| GH89 | 514 | 887 | 0 |
| WLMVPFFRCMLHNFAGNIEMYGVLDAIASGPVEARTSENSTMVGVGMSMEGIEQNPVVYDLMSEMAFQHDKLDVKLWLDVYPMRRYGSSDASIRDAWSIL YHTVYNCTDGAYNKNRDVIVAFPDVDPFSISLEDAYMSGKHQLMSEAVIRRVPLRDSTEAYDRPHIWYSTSDVIHALGLFLASGDDLAESNTYRYDLIDL TRQALAKYANQVFMKIIENYRMNDVHGVTFYSKHFLDLVTDMDTLLACHEGFLLGPWLESSKQHAQTQEQKIQFEWNARTQITMWFDNTEEEASLLRDYG NKYWSGLLKDYYGPRAAVYFKHLLESLENGVGFQLKAWRREWIKLTNDWQKSVKIFPVRSTGNALDTSRWLYEK | |||
| Full Sequence |
|---|
| Protein Sequence Length: 898 Download |
| MIPEVWGSNP ASITELLISL KTGLPNHFFA SVSWAQLVKP LGDDTRGLGF ESRQHYRVID 60 LIKNRLTKPF ANELVAQCAN SSTIGVNHIS QILETHDRER APNSVQVAAS HGVLSRLLPS 120 HSSSFEFQII SKEQCGGKSC FIISNHPSFG INGAPEIRIG GSSGVELSAG LHWYLKNWCG 180 AHISWDKTGG AQLFSVPKPG FLPLVQASGV LIQRPVPWNY YQNAVASSYT FAWWDWERWE 240 KEIDWMAVQG INLPLAFTGQ EAIWQKVFQI FNISSSELND FFGGPAFLAW SRMGNLHGWG 300 GPLPQSWFDQ QLSLQKKILG RMYELGMTPV LPAFSGNVPA ALKSIYPSAK IARLGNWFTV 360 RGDTRWCCTY LLDAMDPLFV EIGKAFVEME ITEYGRSSHI YNCDTFDENT PPVDDPEYIS 420 SLGAAIYRAM QSGDVDAVWL MQGWLFSYDP FWRPEQMKAL LHSVPVGKLV VLDLFAEVKP 480 LWITSEQFYG VPYIWKVAID IANLKWSLSF VSDWLMVPFF RCMLHNFAGN IEMYGVLDAI 540 ASGPVEARTS ENSTMVGVGM SMEGIEQNPV VYDLMSEMAF QHDKLDVKLW LDVYPMRRYG 600 SSDASIRDAW SILYHTVYNC TDGAYNKNRD VIVAFPDVDP FSISLEDAYM SGKHQLMSEA 660 VIRRVPLRDS TEAYDRPHIW YSTSDVIHAL GLFLASGDDL AESNTYRYDL IDLTRQALAK 720 YANQVFMKII ENYRMNDVHG VTFYSKHFLD LVTDMDTLLA CHEGFLLGPW LESSKQHAQT 780 QEQKIQFEWN ARTQITMWFD NTEEEASLLR DYGNKYWSGL LKDYYGPRAA VYFKHLLESL 840 ENGVGFQLKA WRREWIKLTN DWQKSVKIFP VRSTGNALDT SRWLYEKYLR KPDALDQ* 900 |
| Functional Domains Download unfiltered results here | ||||||||
|---|---|---|---|---|---|---|---|---|
 | ||||||||
| Cdd ID | Domain | E-Value | Start | End | Length | Domain Description | ||
| pfam12971 | NAGLU_N | 1.0e-23 | 106 | 205 | 100 | + Alpha-N-acetylglucosaminidase (NAGLU) N-terminal domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This N-terminal domain has an alpha-beta fold. | ||
| pfam12972 | NAGLU_C | 7.0e-94 | 590 | 888 | 299 | + Alpha-N-acetylglucosaminidase (NAGLU) C-terminal domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This C-terminal domain has an all alpha helical fold. | ||
| pfam05089 | NAGLU | 4.0e-180 | 220 | 584 | 365 | + Alpha-N-acetylglucosaminidase (NAGLU) tim-barrel domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This central domain has a tim barrel fold. | ||
| Annotations - NR Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| EMBL | CBI15090.1 | 0 | 79 | 896 | 21 | 845 | unnamed protein product [Vitis vinifera] |
| GenBank | EEC78143.1 | 0 | 88 | 895 | 32 | 813 | hypothetical protein OsI_17702 [Oryza sativa Indica Group] |
| RefSeq | XP_002280399.1 | 0 | 79 | 896 | 21 | 812 | PREDICTED: hypothetical protein [Vitis vinifera] |
| RefSeq | XP_002318632.1 | 0 | 81 | 897 | 26 | 812 | predicted protein [Populus trichocarpa] |
| RefSeq | XP_002511461.1 | 0 | 79 | 894 | 20 | 805 | alpha-n-acetylglucosaminidase, putative [Ricinus communis] |
| Annotations - PDB Download unfiltered results here | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
| PDB | 2vcc_A | 0 | 125 | 878 | 190 | 866 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 Complexed With Udp-Glucose |
| PDB | 2vcb_A | 0 | 125 | 878 | 190 | 866 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 Complexed With Udp-Glucose |
| PDB | 2vca_A | 0 | 125 | 878 | 190 | 866 | A Chain A, Crystal Structure Of Medicago Truncatula Ugt71g1 Complexed With Udp-Glucose |
| PDB | 2vc9_A | 0 | 125 | 878 | 190 | 866 | A Chain A, Family 89 Glycoside Hydrolase From Clostridium Perfringens In Complex With 2-Acetamido-1,2-Dideoxynojirmycin |
| PDB | 4a4a_A | 0 | 125 | 878 | 213 | 889 | A Chain A, Cpgh89 (E483q, E601q), From Clostridium Perfringens, In Complex With Its Substrate Glcnac-Alpha-1,4-Galactose |
| EST Download unfiltered results here | ||||
|---|---|---|---|---|
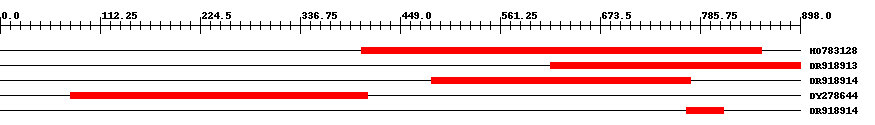 | ||||
| Hit | Length | Start | End | EValue |
| HO783128 | 450 | 406 | 855 | 0 |
| DR918913 | 281 | 618 | 898 | 0 |
| DR918914 | 292 | 484 | 775 | 0 |
| DY278644 | 334 | 79 | 412 | 0 |
| DR918914 | 42 | 771 | 812 | 5e-16 |
| Sequence Alignments (This image is cropped. Click for full image.) |
|---|
 |